Bacterial Modes of Action for Enhancing of Plant Growth
Received: 11-Jul-2016 / Accepted Date: 26-Jul-2016 / Published Date: 02-Aug-2016 DOI: 10.4172/2155-952X.1000236
Abstract
The greatest issue affecting the sustainability of broad acre cropping both environmentally and economically is the requirement of fertilizers. These are based on mined phosphorous or other mineral ores, ammonia produced through the Harbour-Bosch process and industrially manufactured potash. As global demand for fertilizers increases, the costs associated with the production for each of these major nutrients increases. Biofertilizers such as plant growth promoting bacteria (PGPB) are a possible biotechnology that could alleviate the need for addition of increasing amounts of fertilizers. These bacteria naturally occur in soils and aggressively colonize around plant roots and have been shown to have plant growth promoting effects. PGPB are known to influence plant growth by various direct and indirect mechanisms; while some can affect plant physiology directly by mimicking synthesis of plant hormones, others increase mineral availability and nitrogen content in soil. Here we review the previously characterized modes of action for enhancement of plant growth by PGPB such as nitrogen fixation, nutrient solubilization and production of auxins and enzymes, as well as discussing more recent proposed modes of action such as secondary metabolites.
Keywords: Plant growth promoting bacteria (PGPB), Biofertilizer, Rhizosphere nitrogen fixation, Siderophores, Secondary metabolites
Introduction
Globally, agriculture relies on supplementing cropped soils with macro and micronutrients sourced from mined ores or industrially produced through energy intensive processes. The major outcome of supplementing crops with these fertilizers is a consistent yield, however, rising costs associated with producing these fertilizers is tipped to reach a critical point in the next 20 years, where the costs will exceed the pay off, in terms of yield value [1,2]. This is particularly the case for phosphorous based fertilizers for example, which are derived from mined phosphorous ore, but will also become an increasing issue for other essential micronutrients: including zinc, cobalt, magnesium, iron and manganese, which are also sourced from ore deposits.
The main source of commercially available phosphate is derived from phosphate rock. This is a finite resource, and both its acquisition through mining and local depletion of this resource is predicted to have dire impacts on the natural environment. Aside from environmental impacts associated with the mining of phosphate rock, there are large economic costs, which are set to rise as phosphate rock deposits become scarce. It is estimated that world phosphate rock deposits will be depleted within the next 50-100 years [1].
The global price of phosphate rock has risen by over 700% since 2007 during a 14 month period [1-3], and its price is expected increase in the next significantly 20 years (along with the costs of ammonia and potash) [1]. While demand continues to increase, the cost of mining phosphate rock is increasing due to declining quality and greater expense of extraction, refinement, and environmental management [1,4]. The increases in phosphate and the other macro-nutrients costs will inevitably drive increases in farming input prices, resulting in parallel increases in the cost of food production worldwide.
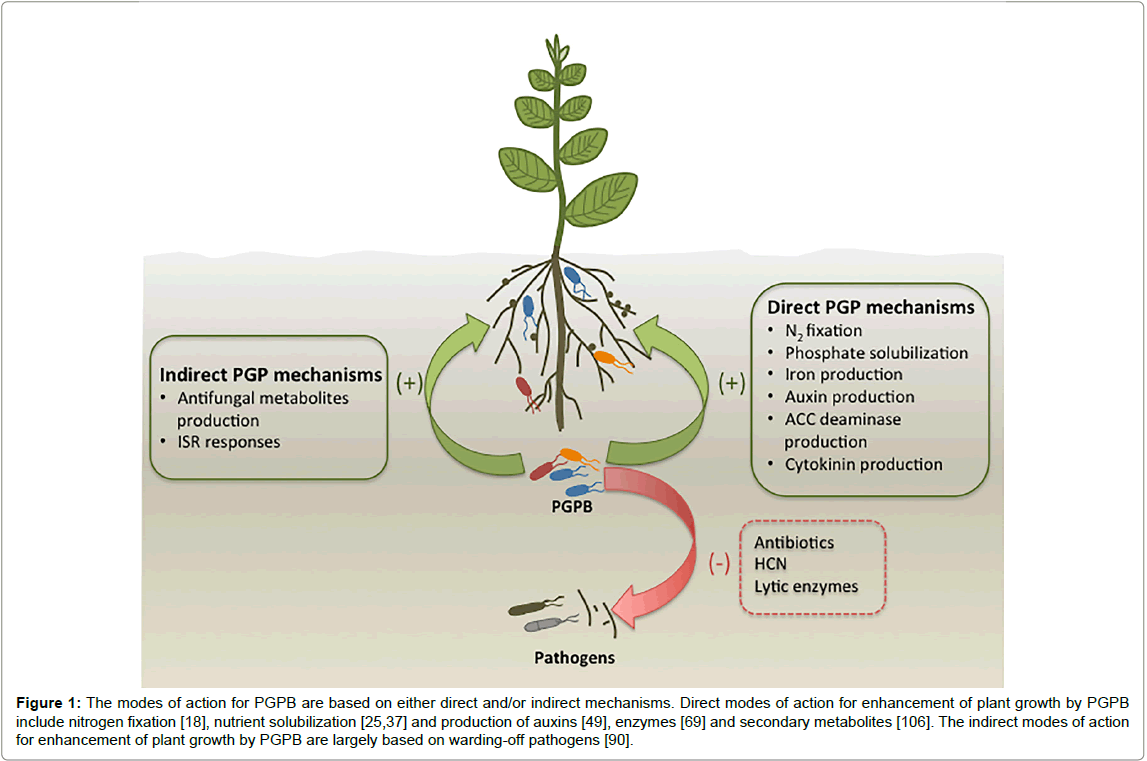
Beneficial microbes that exist in soils naturally are known as plant growth promoting bacteria (PGPB). The potential of PGPB to reduce dependence on high levels of fertilizer inputs has gained significant increase in interest over recent years [5-7]. Pseudomonas, Klebsiella, Enterobacter, Azospirillum, Bacillus, Alcaligenes and Arthrobacter are genera that have been reported to enhance plant growth [8-10] and these PGPB strains are capable of enhancing plant growth either directly or indirectly (Figure 1) [5]. The direct promotion of plant growth is based on the bacterial production of phytohormones, enzymes, siderophores or amino acids [5]. The indirect enhancement of plant growth promotion by bacteria is largely through the restriction of infection and prevalence of deleterious pathogenic organisms through the bacterial production of antagonistic substances (Figure 1) [5].
Figure 1: The modes of action for PGPB are based on either direct and/or indirect mechanisms. Direct modes of action for enhancement of plant growth by PGPB include nitrogen fixation [18], nutrient solubilization [25,37] and production of auxins [49], enzymes [69] and secondary metabolites [106]. The indirect modes of action for enhancement of plant growth by PGPB are largely based on warding-off pathogens [90].
Plant-Microbe Interactions in the Rhizosphere
The thin layer of soil surrounding plant roots is known as the rhizosphere, and is an extremely important area for plant-microbe interactions. It is approximately one millimeter wide and characterized by high levels of biological and chemical activities and is comprised of plant, fungi, bacteria and soil constituents [11]. The rhizosphere can contain over 1010 microbial cells per gram of root [11] and sustain more than 30,000 species of bacteria [12]. The microbial populations benefit from root exudates including: vitamins, sugars, proteins, carbohydrates, organic acids, amino acids and mucilage [13]. These root exudates are also capable of modifying physical and biochemical properties of the rhizosphere by acting as messengers between plant and microbe [13]. For instance, benzoxazinoids found in the root exudates of maize attract Pseudomonas putida, which is a competitive colonizer of the maize rhizosphere with plant beneficial traits [14].
Biological Nitrogen Fixation (BNF)
Nitrogen is one of the three macronutrients required for high crop yields. Three quarters of our atmosphere consists of nitrogen gas (N2) and elemental nitrogen must be transformed to usable forms before it is available for plant uptake. Nitrogen is the major component of chlorophyll, which plays an important role in photosynthesis. It is also a building material of proteins and a major component of DNA and RNA in the form of the nitrogenous bases. In terms of nutrient demand, plants require more nitrogen than any other nutrient, however, there is a very limited amount of nitrogen available in soil due to regular nitrogen loss.
Nitrogen can be lost from the rhizosphere through the following processes:
Denitrification- Oxidative reduction of soil nitrates to atmospheric nitrogen by heterotrophic facultative anaerobic bacteria through a serious of gaseous nitrogen oxide intermediates [15].
Volatilization- Loss of the organic form of nitrogen as urea (originated from animal manure, fertilizers and plant materials), which is converted into gaseous ammonia to the atmosphere [16]
Leaching- Once the nitrogen has been converted to nitrate, the excess soil nitrates dissolved in water can move below the root zone under certain conditions. Generally, leaching takes place in sandy soils with low water holding capacity where water penetrates quickly. The amount of leaching is also dependent on subsoil moisture recharge [17]. If subsoil moisture levels are not recharged, water is more likely to be held in soil, therefore reduces the probability of leaching [17].
Certain microbes are able to convert atmospheric nitrogen into utilizable forms of nitrogen for both themselves and plants via biological fixation process [18]. These nitrogen-fixing microorganisms can be divided into two main categories, symbiotic and free-living. It is well documented that biological nitrogen fixation (BNF) is facilitated by an multimeric enzyme called nitrogenase, which consists of conserved proteins, iron containing dinitrogenase reductase and molybdenum iron dinitrogenase [19]. These enzymes are irreversibly inhibited by molecular oxygen, therefore nitrogen-fixing bacteria have developed evolutionary adaptive mechanisms to limit oxygen exposure via leghemoglobin [20]. The nitrogen fixation capacity of bacteria is highly dependent on moisture content, oxygen concentration and supply of organic substrates in soil [19].
Of all the bacterial species that have the ability to fix atmospheric nitrogen [21], rhizobia are the best described for providing nitrogen to plants, and are well known for their ability to form nodules on roots of legume plants. The success of rhizobium in providing nitrogen to their legume hosts has led to the commercialization of rhizobium inoculants. However, the efficiency of these bacterial inoculants can be greatly affected by rhizosphere nutrient conditions.
Symbiotic nitrogen-fixing bacteria reduce atmospheric nitrogen within the root nodules to ammonia, which is then being used by the host plant [22]. This symbiosis process allows the bacteria to have an exclusive niche and carbon source and in return, the plant will obtain nitrogen. Similar relationships can be observed in aerobic Azotobacter and anaerobic Clostridia with higher plants such as Azolla [22].
Enhancement of Phosphate Availability to Plants by Bacteria
Phosphorus (P) is a crucial element for survival and prosperity of most organisms including plants and is the second most important plant growth-limiting nutrient after nitrogen. The majority of phosphorus is in its insoluble form, whilst the plants can only absorb phosphorus when it is bonded with oxygen as in monobasic (H2PO4-) and dibasic forms (HPO42-) [23]. A low abundance of phosphorus is typical in many agricultural soils, and where phosphates are often complexed to soil constituents, making them unavailable to many organisms [24]. To overcome plant growth limiting phosphate deficiency, phosphate dense fertilizers are applied to crops regularly. However, plants can only absorb limited amount of phosphates and the rest is rapidly converted into insoluble P. There is also an extensive loss of phosphates in agricultural lands via run off and much of the phosphate ends up in water reservoirs.
Phosphate is essential for the optimum growth of most bacteria and has a central role in many metabolic and energy producing pathways. Microorganisms associated with hydrolyzing organic and inorganic phosphates are known as phosphate-solubilizing bacteria (PSB). These organisms are known to solubilize P from substrates and make it available to plants, and hence are a possible alternative to phosphate rich fertilizers. Bacteria of the genera Azotobacter, Bacillus, Beijerinckia, Burkholderia, Enterobacter, Erwinia, Flavobacterium, Microbacterium, Pseudomonas, Rhizobium and Serratia are reported as the most effective PSB [25].
Environmental factors such as temperature, concentration of iron, carbon and nitrogen sources can impact on P solubilizing ability of bacteria. PSB are able to solubilize insoluble forms of P such as aluminum phosphate (Al3PO4), iron phosphate (Fe3PO4) and tricalcium phosphate (Ca3(PO4)2) [26]. Typically, inorganic phosphate solubilizaton is initiated as a consequence of the actions of low molecular organic acids such as gluconic and citric acid [26]. Organic P compounds undergo mineralization and the resulting P will be readily available for the plants to absorb [27,28]. This mineralization process is facilitated by enzymes secreted by soil microbes such as phosphatases [29] and phytases [30]. Considering critical impacts of such enzymes in dissolution of complex compounds into usable P, it is highly desirable to develop the bacterial inoculants with high phosphatase and phytase activity to overcome P limiting soils.
Bacterial Siderophore Production
Iron is a vital nutrient for proper plant development. Since it is a cofactor of many metabolic pathways, its deficiency may lead to disruption of many processes including respiration or photosynthesis. In aqueous environments, iron exists as Fe3+ and Fe2+, which form insoluble oxides and hydroxides. However, both plants and microorganisms are unable to metabolize insoluble iron oxides [31].
Plants mainly acquire Fe from the rhizosphere and have developed two strategies to acquire iron. The first strategy involves acidification of the rhizosphere, followed by the reduction of Fe3+ ions by membranebound Fe3+-chelate reductase, and subsequent uptake of Fe2+ into root cells [32]. The second strategy involves plants secreting low molecular weight phytosiderophores in order to solubilize and bind iron, which is then transported into root cells via membrane proteins [33,34]. These strategies are often not efficient enough to meet the needs of the plants growing, particularly in calcareous and alkaline soils. Therefore, providing plants with accessible forms of iron is often necessary, particularly for intensively cropped soils [35].
The application of PGPB may be one strategy to increase soil iron availability in the rhizosphere. Bacteria are able to synthesize low molecular weight siderophores, which have the ability to scavenge Fe3+ from the environment [36]. Siderophores have high affinity ligands that are able to pair with ferric ions. They have a strong capacity to chelate ferric ions allowing their solubilization and extraction from most mineral or organic complexes [37].
Siderophores are small peptide metabolic molecules with functional groups, which can provide a set of high affinity ligands to equalize ferric ions. Bacterial siderophores have been classified into three main categories according to their structural features, type of ligands, and their iron coordinating functional groups. They are namely carboxylates, hydroxamates and catecholates [38]. Hydroxamate siderophores have a 1:1 (metal-EDTA complexes) stability constant with Fe3+ that nears that of the Fe3+-EDTA complex (1030), whereas catecholates and carboxylates siderophores can form similar complexes with stability constants near that of Fe3+-EDDHA (1040) [39].
The ability to produce siderophores plays a central role in various microorganisms in regards to plant growth promotion. Bacterial siderophores from Chryseobacterium spp. C138 isolated from the rhizosphere of Oryza sativa are effective in supplying Fe to iron-starved tomato plants when delivered to the roots [40]. Supplementation of fluorescent Pseudomonas strains in maize seeds was demonstrated to significantly increase germination percentage and plant growth [41]. Under low iron conditions, co-inoculation of maize with siderophore producing plant growth promoting rhizobacteria, resulted in larger shoot and root lengths and higher dry weights, in comparison with uninoculated plants suggesting application of these bacteria could increase crop productivity in calcareous soils [41].
Studies have shown that plants absorbed radiolabelled iron from siderophores previously exposed to radiolabelled iron [42]. This can be observed in Fe-pyoverdine complex synthesized by Pseudomonas fluorescence C7, which was then taken up by Arabidopsis thaliana plants in order to fulfill their iron requirements [42].
Bacterial Auxin Production
Auxins are plant hormones that are essential for plant development. They have a fundamental role in coordination of many growth and physiological processes in the plant's life. Indole-3-acetic acid (IAA) is the most extensively studied plant hormone and it is a carboxylic acid with a carboxyl group attached through a methylene group to the 3rd carbon of the indole ring [43]. IAA affects plant nutrition and development through altering cell division, extension and differentiation by increasing the rate of xylene and root development [44].
Plant growth promoting bacteria exhibit a variety of characteristics responsible for influencing plant growth, including the production of IAA [45,46]. Indole-3-acetic acid in rhizobacteria helps to loosen plant cell walls, which may facilitate rhizobacteria to absorb various substances secreted by roots [44]. Several studies have suggested that elevated auxin levels, including IAA in host plants, are required for nodule formation [45,47-49]. This was observed when low IAA producing Bradyrhizobium elkanii mutants were shown to result in fewer nodules in soybean roots than the wild-type strain [45]. Coinoculation of low IAA producing mutant of Rhizobium sp. NGR234 with soy bean shows that the IAA content in nodules is significantly lower than in nodules induced by the wild-type strain [50]. This supports the concept that some elements of the IAA established in plant nodules is of prokaryotic ancestry and IAA facilitates nodulation.
The amino acid tryptophan is vital for regulating the levels of IAA synthesized by bacteria [51], while anthranilate acts as a precursor for tryptophan [52]. By this mechanism, IAA biosynthesis is fine-tuned because tryptophan inhibits anthranilate formation by a negative feedback regulation on the anthranilate synthase, resulting in an indirect induction of IAA production [52]. It has been shown that the supplementation of tryptophan could increase IAA production in most rhizobacteria [50].
IAA produced by rhizobacteria may be involved in various plant-bacterial interactions. Most rhizobium strains that have been investigated for plant growth promotion have been found to produce IAA [53]. A recent study investigating the role of IAA synthesized by Pseudomonas putida GR12-2 has confirmed that it promoted canola root development by 35-50% over the roots from seeds treated with IAA deficient mutant, and the roots from uninoculated seeds [46].
Reduction of Plant Ethylene Levels by Bacterially Produced 1-Aminocyclopropane-1-Carboxylate (ACC) Deaminase
Ethylene is an essential metabolite for normal growth and development of plants [54]. This growth hormone is produced by all plants (particularly in rapidly growing cells) and is capable of inducing various physiological changes [55]. Ethylene has a very low solubility in water, however, it can efficiently diffuse through cell membranes. Plant cells in developing seedlings produce elevated amounts of ethylene to increase cell numbers [56]. However, as shoots expand, phytohormones generate a signal to limit the production of ethylene in order to allow leaf expansion [57]. In pea shoots, it has been shown that when they meet an obstacle during their rapid growth stage, reduced levels of ethylene are produced. This allows lateral expansion of the stem resulting in radial enlargement [58]. It has also been identified that ethylene is responsible for thicker and steadier tree trunks and branches [58]. Moreover, ethylene affects fruit ripening process and elevated ethylene levels in fully ripen fruits resulted in a climacteric event just before the seed dispersal [59].
Apart from being a plant growth regulator, ethylene has a vital role as a growth inhibitor [60]. Levels of ethylene in plants increase under stressful circumstances such as drought, presence of heavy metals, extreme temperatures, radiation, high salt environments and wounding pathogenicity [61]. In such situations, plants will initiate survival strategies, which may result in low crop yields [61]. It has been shown that plants exposed to adverse conditions quickly respond by producing a small peak of ethylene due to the activation of a protective mechanism by plants [62]. If the severe conditions persist for a few days a second peak of ethylene arises. After the second ethylene peak, processes such as senescence, chlorosis and abscission are induced [62].
ACC deaminase is an enzyme produced by PGPB, which can decrease ethylene levels, thus increase stress (salt and drought) tolerance in plants [63]. There is a wide range of bacteria that have been identified as ACC deaminase positive including: Pseudomonas, Ralstonia, Serratia, Rhizobium, Acinetobacter, Agrobacterium, Achromobacter, Alcaligenes and Bacillus [64-67]. These bacteria are capable of taking up the ethylene precursor ACC from plants and converting it into 2-oxobutanoate and ammonia [68]. By decreasing ACC levels in plants, ACC deaminase producing bacteria limit accumulation of high ethylene concentrations in plants.
Environmental stressors such as heavy metals in soil, radiation, extreme temperatures, insect predation, high light intensity and high salt concentration are alleviated by the production of ACC deaminase [69]. Thus, co-inoculation of ACC deaminase producing bacteria with plants subjected to a wide range of biotic and abiotic stressors, can result in enhanced plant tolerance [69].
Enhancement of Plant Growth by Bacterially Produced Cytokinins
Cytokinins are organic plant growth hormones that have the ability to promote cell division (cytokinesis) in plant root and shoots. Naturally occurring cytokinins are adenine derivatives and they carry either an isoprene derived side chain or an aromatic side chain at the N6 terminus [70]. They are called isoprenoid, cytokinins or aromatic cytokinins, respectively [71,72]. Isoprenoid cytokinins are commonly found in a range of plant species, however, there is no clear evidence of the common existence of aromatic cytokinins in plants [70]. Cytokinins influence cell division by stimulating the production of proteins required for mitosis and are produced in the meristem where stem cells self-renew and produce daughter cells that separate and give rise to different organ structures [73]. Once synthesized in roots, they travel up the xylem to the other parts of the plant such as leaves, developing fruits and seeds [73].
Seven different genes (IPT: adenosine phosphateisopentenyltransferase) have been identified as potential cytokinin producers in Arabidopsis (altPT1 and altPT3 to altPT8) [74-76]. AltPT1, AltPT3, AltPT5 and AltPT8, are expressed in plastids and produce cytokinins with dimethylallyl diphosphate (DMAPP) originated from MEP (Methylerythritol phosphate) pathway [77]. AltPT4 and AltPT7 are located in the cytosol and mitochondria respectively, and they produce cytokinins with DMAPP derived from MVA (Mevolonate) pathway [77]. Inoculation of lettuce plants with cytokinin producing bacterium, Bacillus subtilis increased the cytokinin content of both shoot and roots [78]. Accumulation of cytokinin, zeatin riboside in inoculated lettuce plants was associated with an overall increase in both root and shoot weights [78].
Cytokinins have recently been found to play a crucial role in resistance to plant pathogens. Plant-derived cytokinins promote resistance against the non-cytokinin producing pathogen, Pseudomonas syringae by modulating the defence signalling in Arabidopsis [79]. In Nicotiana tobocum, cytokinins enhanced overall plant resistance against the virulent hemibiotrophic pathogen, P. syringae [80]. This cytokinin-mediated plant resistance is associated with elevated levels of bacteriocidal activities and increased amounts of antimicrobial phytoalexines [80]. In the context of plant biological control via microbes, cytokinins produced by P. fluorescens G20-18 have negative influence on the pathogenic activity of P. syringae in A. thaliana [81].
Enhancement of Plant Growth by Bacterially Produced Secondary Metabolites
In the past, secondary metabolites were considered as elements with a low molecular mass that are not the end products of primary metabolic pathways [82,83]. It was assumed that secondary metabolites were not vital to normal bacterial function, hence had no influence on growth and development of microorganisms. Contrary to this, recent studies have shown that secondary metabolites are fundamental for growing cells as regulators of cellular differentiation processes and also as cellular inhibitors against competing bacteria [84-86].
A vast range of secondary metabolites have been discovered and despite their greater divergence, most secondary metabolites have the same precursors branched into various compounds that result in secondary metabolites [86]. Depending on their structural significance, secondary metabolites have distinct features that reflect their biological activity. Apart from their involvement in medicine, these bioactive compounds have demonstrated compelling advantages in agricultural and industrial applications [85].
Antibiotics produced by bacteria are some of the best known secondary metabolites and approximately 12,000 antibiotics have been identified [87]. The majority of bacterial antibiotics are produced by filamentous bacteria of the genus, Actinomyces [86]. Parallel to the screening for new antibiotics, research efforts have been targeted towards identifying bioactive compounds with other biological activities such as herbicides, immunosuppressant agents, insecticides, plant growth promoter/inhibitors, enzyme inhibitors and antihelmintics.
Functional relevance of secondary metabolites produced by bacteria has yet to be determined in terms of their effects on the soil bacterial ecology. It has been found that microbial bioactive agents such as antibiotics are not only beneficial to hosts against various microbial competitors, but also against other organisms including insects, parasites and plants [86,88]. Most potential secondary metabolites are present in very low amounts in the rhizosphere.
Bacterial Antibiotic Production Enhances Plant Growth
The production of antifungal antibiotics depends on biocontrol ability and the degree of bacterial colonization in plant roots [89]. Bacterially produced antibiotics are mainly low molecular weight organic compounds with heterogeneous groups, which can have deleterious effect on pathogenic of microorganisms. PGPB can act as antagonistic agents against plant pathogens by producing one or more antibiotics. There are six classes of biocontrol antibiotic agents depending on their mode of action: phenazines, phloroglucinols, pyrrolnitrin, pyoluteorin, hydrogen cyanide (HCN) and cyclic lipopeptides [90].
Phenazines
Phenazines are the pigments produced by eubacteria [91]. Production of phenazines varies in different organisms and these phenazines are released at high levels (milligrams to gram per liter) during bacterial growth in vivo. Phenzines absorb in both the UV and visible range that alter due to arrangement of the heterocyclic ring [92]. Functional groups present in heterocyclic ring are subjected to the variations in their biological activity [92].
Phenazine production has been demonstrated in various experiments and is common among bacteria with high guanine and cytosine (G+C) content [93]. Fluorescent pseudomonads including P. fluorescence and P. chlororaphis are regarded as being the highest producers of phenazines [94]. Phenazine producing fluorescent pseudomonads typically produces two or more phenazines except P. fluorescence, which produces only phenazine-1-carboxylic acid (PCA). In addition to PCA, P. chlororaphis produces phenazine-1- carboxamide (PCN), whereas P. aeruginosa produces 5-N-methyl- 1-hydroxyphenazine (PYO) [94]. Not only PYO, P. aeruginosa is also capable of synthesizing PCN, aeruginosin A (5-methyl-7-amino-1- carboxymethylphenazinium betaine) and aeruginosin B (5-methyl-7- amino-1-carboxy-3-sulfophenazinium betaine) [94]. A microorganism that belongs to Enterobacteriaceae is Pantoea agglomerans Eh 1087, which produces alanylgriseoluteic acid (AGA). Similarly, phenazines have been identified in marine bacteria known as Pelagiobacter variabilis and Vibrio sp. SANK73794 [95].
Phenazine is a bioactive metabolite that inhibits the growth of microorganisms. This has been confirmed in field grown wheat, where a concentration of 100 nM of PCA inhibited growth of certain Grampositive bacteria and fungi [96]. Further investigation revealed that PCA produced by P. fluorescens in the wheat rhizosphere was linked to the inhibition of the wheat pathogen, Gaeumannomyces graminis var. tritici [97,98]. Similar studies of the plant pathogenic fungus, Rhizoctonia spp. in wheat revealed that the application of phenazine producing Pseudomonas spp. restricted Rhizoctonia caused root rot [99].
Phenazine also enhances the survival of bacteria in anaerobic conditions. In such conditions, endogenous phenazines enhance the survival of P. aeruginosa by facilitating extracellular electron transfer [100]. Moreover, PCA has the ability to increase iron bioavailability via reducing Fe3+ to Fe2+ [101]. This is evident when a mutant form of P. aeruginosa that is unable to form siderophores can enhance formation of biofilms in the presence of PCA, making iron more bioavailable in the environment [101].
Phloroglucinol
Phloroglucinol is a benzenetriol, mainly used in pharmaceutical production of Flopropione (antispasmodic agent) [102]. Phloroglucinol is synthesized commercially via a number of processes, however selective trinitration of benzene is the widely used technique in the world. Phloroglucinols are naturally found in certain plant species and are also produced by microorganisms [94]. Pseudomonas fluorescens produces phloroglucinol with a type III polyketide synthase [103]. Synthesis begins via the condensation of three malonyl-CoAs which are coenzyme A derivatives of malonic acid [103]. While de-carboxylation of 3,5-diketoheptanedioate, cyclization of the activated product leads to the formation of phloroglucinol [103].
The 2,4-diacetylphloroglucinol (DAPG) is the widely studied phloroglucinol produced by Pseduomonads. It can cause membrane and zoospore damage in Pythium spp. This antibiotic acts as an inhibitor to aldose reductase, an enzyme involved in metabolism of glucose to fructose [104]. DAPG is not only a direct antagonist of plant pathogen but also it is resistive in Arabidopsis thaliana against Peronospora parasitica, a water mold [105]. Although, DAPG exhibits antifungal activity, it also acts as a plant growth stimulator. DAPG produced by P. fluorescens isolates containing the phID gene that can stimulate lateral root formation in tomato seedlings by inhibiting primary root development [106]. This indicates that DAPG can change the root architecture by merging with an auxin dependent signaling pathway [106].
Enhancement of Plant Growth by Bacterially Produced Antifungal and Antibiotic Products
Pyrrolnitrin (3-chloro-4-(3-chloro-2-nitrophenyl) pyrrole) is an antifungal antibiotic produced by many fluorescent and non-fluorescent bacterial strains, and it is initially isolated from Burkholderia pyrrocina [107]. Pyrrolnitrin inhibits the growth of fungi, yeast and bacteria by reacting with phospholipid components, which leads to burst cellular membrane [108]. Predominantly, this antibiotic is used to restrain the growth of dermatophytic fungi, such as Trichophyton species [109]. Studies performed on P. aureofaciens indicated pyrrolnitrin has a relatively closer arrangement to tryptophan (IAA synthesis precursor) [110]. Further analysis revealed that the addition of D-tryptophan in growth medium can increase the pyrrolnitrin production, whereas L-tryptophan had no significant effect on the pyrrolnitrin production [110].
A type of pyrrolnitrin produced by P. fluorescens BL15 strain has been demonstrated to inhibit the damage caused by Rhizoctonia solani during damping-off of cotton plants [111]. Pseudomonas fluorescens BL15 has been shown to produce other antifungal metabolites including HCN, chitinase and 2-hexyl-5-propyl-resorcinol [112]. Pyrrolnitrin (approximately 10 μg/mL) is capable of inhibiting the growth of Saccharomyces cerevisiae, Penicillium atrovenetum and P. oxalicum [113]. It affects the terminal electron transport system between succinate or NADH (reduced nicotinamide adenine dinucleotide) and coenzyme Q [113]. At such level, pyrrolnitrin inhibits endogenous and exogenous respiration immediately after its addition to the system.
Conclusion
It is clear that microbes have the potential to enhance plant growth through various mechanisms and may help to reduce chemical fertilizer inputs. Whilst some modes of actions such as auxin, enzyme production and nutrient (phosphate and iron) solubilization have been well studied, there are still many areas that require further investigation. One promising target is the area of secondary metabolite production by PGPB as many of these metabolites have not been well characterized and thus further understanding is required for identification of modes of action for plant growth promoting strains. The nature of the these molecules and the fact that they are often produced in very small amounts, requires the use of modern separation and analytical techniques such as high performance liquid chromatography and gas/ liquid chromatography–mass spectrometry.
References
- Cordell DJ, Drangert J, White S (2009) Thestory of phosphorus: Global food security and food for thought. Global Environmental Change 19: 292-305.
- Shepherd JG, Kleemann R, Bahri-Esfahani J, Hudek L, Suriyagoda L, et al. (2016) The future of phosphorus in our hands. NutrCyclAgroecosyst 104: 281-287.
- Schnitkey G (2014) Controlling costs with lower crop revenues: Machinery costs. Department of Agricultural and Consumer Economics, University of Illinois, Champaign, USA.
- Cordell DJ, White S (2013) Sustainable phosphorus measures: Strategies and technologies for achieving Phosphorus security. Agronomy 3: 86-116.
- Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41: 109-117.
- Lucy M, Reed E, Glick BR (2004) Applications of free living plant growth-promoting rhizobacteria. Antonie van Leeuwenhoek 86: 1-25.
- Vessey JK (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255: 571-586.
- Kloepper JW, Lifshitz R, Zablotowicz RM (1989) Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol 7: 39-43.
- Okon Y, Labandera-Gonzalez CA (1994) Agronomical applications of Azospirillum. In improving plant productivity with rhizospherebacteria. Soil BiolBiochem 26: 1591-1601.
- Joseph B, Patra RR, Lawrence R (2007) Characterization of plant growth promoting rhizobacteriaassoicated with chickpea. Int J Plant Prod 1: 141-152.
- Egamberdieva D, Kamilova F, Validov S, Gafurova L, Kucharova Z, et al. (2008) High incidence of plant growth stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ Microbiol 10: 1-9.
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, et al. (2011) Deciphering the rhizospheremicrobiome for disease-suppressive bacteria. Science 332: 1097-1100.
- Brimecombe MJ, De Leij FA, Lynch JM (2000) The effect of root exudates on rhizosphere microbial populations. Marcel Dekker Inc, New York, USA.
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attractPseudomonas putida to the rhizosphere. Plos one 7: 1-11.
- Burford J, Bremner J (1975) Relationships between the denitrification capacities of soils and total, water-soluble and readily decomposable soil organic matter. Soil BiolBiochem 7: 389-394.
- Terman G (1979) Volatilization losses of nitrogen as ammonia. AdvAgron 31: 189-223.
- Kløve B, Ala-Aho P, Bertrand G, Gurdak JJ, Kupfersberger H, et al. (2014) Climate change impacts on groundwater and dependent ecosystems. J Hydrol 518, Part B: 250-266.
- Kim J, Rees DC (1994) Nitrogenase and biological nitrogen fixation. Biochemistry 33: 389-397.
- Berman-Frank I, Lundgren P, Falkowski P (2003) Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol 154: 157-164.
- Church MJ, Björkman KM, Karl DM, Saito MA, Zehr JP (2008) Regional distributions of nitrogen-fixing bacteria in the Pacific Ocean. LimnolOceanogr 53: 63-77.
- Nghia NH, Gyurjan (1987) Problems and perspectives in establishment of nitrogen-fixing symbioses and endosymbioses. Endocytobiosis Cell Res 4: 131-141.
- Shridhar BS (2012) Reveiw: Nitrogen fixing microorganisms.Int J Microbiol Res 3: 46-52.
- Kruse J, Abraham M, Amelung W, Baum C, Bol R, et al. (2015) Innovative methods in soil phosphorus research: A review. J Plant Nutr Soil Sci 178: 43-88.
- Lambers H, Ahmedi I, Berkowitz O, Dunne C, Finnegan PM, et al. (2013) Phosphorus nutrition of phosphorus-sensitive Australian native plants: threats to plant communities in a global biodiversity hotspot. ConservPhysiol 1: 1-21.
- Bhattacharyya P, Jha D (2012) Plant growth promoting rhizobacteria (PGPR): Emergence in agriculture. World J MicrobiolBiotechnol 28: 1327-1350.
- Song OR, Lee SJ, Lee YS, Lee SC, Kim KK, et al. (2008) Solubilization of insoluble inorganic phosphate by Burkholderiacepacia DA23 isolated from cultivated soil. Braz J Microbiol 39: 151-156.
- Tarafdar JC, Claassen N (1988) Organic phosphorus compounds as a phosphorus source for higher plants through the activity of phosphatases produced by plant roots and microorganisms. BiolFertil Soils 5: 308-312.
- Rodríguez H, Fraga R, Gonzalez T, Bashan Y (2006) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287: 15-21.
- Tarafdar JC, Rao AV, Bala K (1988) Production of phosphatates by fungi isolated from desert soils. Folia Microbiol 33: 453-457.
- Maougal RT, Brauman A, Plassard C, Abadie J, Djekoun A, et al. (2014) Bacterial capacities to mineralize phytate increase in the rhizosphere of nodulated common bean (Phaseolus vulgaris) under P deficiency. Eur J Soil Biol 62: 8-12.
- Rajkumar M, Ae N, Prasad MNV, Freitas H (2010) Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28: 142-149.
- Mori S (1999) Iron acquisition by plants. CurrOpin Plant Biol 2: 250-253.
- Altomare C, Tringovska I (2011) Beneficial soil microorganisms, an ecological alternative for soil fertility management. Springer Dordrecht, Netherlands.
- White P, Karley A, Potassium RH, Mendel R (2010) Cell biology of metals and nutrients. Springer, Berlin, Germany.
- Zuo Y, Zhang F (2011) Soil and crop management strategies to prevent iron deficiency in crops. Plant Soil 339: 83-95.
- Neilands J (1995) Siderophores: Structure and function of microbial iron transport compounds. J BiolChem 270: 26723-26726.
- Sandy M, Butler A (2009) Microbial iron acquisition: Marine and terrestrial siderophores. Chem Rev 109: 4580-4595.
- Crowley D (2006) Microbial siderophores in the plant rhizosphere. Springer, Dordrecht, Netherlands.
- Robert M, Chenu C (1992) Interactions between soil minerals and microorganisms. Marcel Dekker, INC, New York, USA.
- Radzki W, Manero FG, Algar E, García JL, García-Villaraco A, et al. (2013) Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 104: 321-330.
- Sharma A, Johri B (2003) Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS 9 in maize (Zea mays L.) under iron limiting conditions. Microbiol Res 158: 243-248.
- Vansuyt G, Robin A, Briat JF, Curie C, Lemanceau P (2007) Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol Plant Microbe Interact 20: 441-447.
- Thimann KV (1939) Auxins and the inhibition of plant growth. Biol Rev 14: 314-337.
- Glick BR (2012) Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012: 1-15.
- Fukuhara H, Minakawa Y, Akao S, Minamisawa K (1994) The involvement of indole-3-acetic acid produced by Bradyrhizobiumelkanii in nodule formation. Plant Cell Physiol 35: 1261-1265.
- Patten CL, Glick BR (2002) Role of Pseudomonas putidaindoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68: 3795-3801.
- Mathesius U, Schlaman HR, Spaink HP, Of Sautter C, Rolfe BG, et al. (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23-34.
- Pii Y, Crimi M, Cremonese G, Spena A, Pandolfini T (2007) Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol 7: 1-9.
- Hunter WJ (1987) Influence of 5-methyltryptophan-resistant Bradyrhizobiumjaponicum on soybean root nodule indole-3-acetic acid content.Appl Environ Microbiol 53: 1051-1055.
- Spaepen S, Vanderleyden J (2011) Auxin and plant-microbe interactions. CSH PerspectBiol 3: 1-13.
- Zhao Y (2012) Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334-338.
- Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425-448.
- Badenoch-Jones J, Summons RE, Rolfe BG, Letham DS (1984) Phytohormones, rhizobium mutants and nodulation in legumes. IV. Auxin metabolites in pea root nodules. J Plant Growth Regul 3: 23-39.
- Khalid A, Akhtar MJ, Mahmood MH, Arshad M (2006) Effect of substrate-dependent microbial ethylene production on plant growth. Microbiology 75: 231-236.
- Burg SP (1973) Ethylene is plant growth. ProcNatlAcadSci USA 70: 591-597.
- Samimy C (1978) Effect of light on ethylene production and hypocotyl growth of soybean seedlings. Plant Physiol 61: 772-774.
- Lee K, Seo PJ (2015) The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in dopsis. Plant Signal Behav 10: 1-4.
- Taiz L, Rayle D, Eisinger W (1983) Ethylene-induced lateral expansion in etiolated pea stems: The role of Acid secretion. Plant Physiol 73: 413-417.
- Abeles FB, Takeda F (1990) Cellulase activity and ethylene in ripening strawberry and apple fruits. SciHort 42: 269-275.
- Abeles FB, Morgan PW, SaltveitJr ME (1992) Ethylene in plant biology.Academic Press, New York, USA
- Morgan PW, Drew MC (1997) Ethylene and plant responses to stress.Physiol Plant 100: 620-630
- Glick B, Cheng Z, Czarny J, Duan J (2007) Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur J Plant Pathol 119: 329-339.
- Tao JJ, Chen HW, Ma B, Zhang WK, Chen SY, et al. (2015) The role of ethylene in plants under salinity stress. Front Plant Sci 6: 1-12.
- Nadeem SM, Zahir ZA, Naveed M, Arshad M (2007) Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can J Microbiol 53: 1141-1149.
- Nadeem SM, Zahir ZA, Naveed M, Arshad M (2009) Rhizobacteria containing ACC deaminase confer salt tolerance in maize grown on salt-affected fields. Can J Microbiol 55: 1302-1309
- Shaharoona B, Arshad M, Khalid A (2007) Differential response of etiolated pea seedlings to inoculation with rhizobacteria capable of utilizing 1-aminocyclopropane-1-carboxylate or L-methionine. J Microbiol 45: 15-20.
- Shaharoona B, Jamro GM, Zahir ZA, Arshad M, Memon KS (2007) Effectiveness of various Pseudomonas spp. and Burkholderiacaryophylli containing ACC deaminase for improving growth and yield of wheat (Triticumaestivum L.). J MicrobiolBiotechnol 17: 1300-1307.
- Arshad M, Saleem M, Hussain S (2007) Perspectives of bacterial ACC deaminase in phytoremediation. Trends Biotechnol 25: 356-362.
- Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169: 30-39.
- Hwang I, Sakakibara H (2006) Cytokinin biosynthesis and perception. Physiol Plant 126: 528-538.
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant MolBiol 52: 89-118.
- DelloIoio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, et al. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. CurrBiol 17: 678-682.
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyldiphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol 42: 677-685.
- Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylateisopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J BiolChem 276: 26405-26410.
- Sun J, Niu QW, Tarkowski P, Tarkowski P, Zheng B, et al. (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyltransferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131: 167-176.
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, et al. (2004) Distinct isoprenoid origins of cis and trans-zeatinbiosyntheses in Arabidopsis. J BiolChem 279: 14049-14054.
- Arkhipova T, Veselov S, Melentiev A, Martynenko E, Kudoyarova G (2005) Ability of bacterium Bacillussubtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil 272: 201-209.
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, et al. (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19: 284-295.
- Großkinsky DK, Naseem M, Abdelmohsen UR, Plickert N, Engelke T, et al. (2011) Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol 157: 815-830.
- Großkinsky DK, Tafner R, Moreno MV, Stenglein SA, de Salamone IEG, et al. (2016) Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci Rep 6: 23310.
- Vining L (1992) Secondary metabolism, inventive evolution and biochemical diversity-a review. Gene 115: 135-140.
- Yarbrough GG, Taylor DP, Rowlands RT, Crawford MS, Lasure LL (1993) Screening microbial metabolites for new drugs: Theoretical and practical issues. J Antibiot 46: 535-544.
- Stone M, Williams D (1992) On the evolution of functional secondary metabolites (natural products). MolMicrobiol 6: 29-34.
- DemainA (1999) Pharmaceutically active secondary metabolites of microorganisms. ApplMicrobiolBiotechnol 52: 455-463.
- Demain AL, Fang A (2000) The natural functions of secondary metabolites. Springer Berlin, Germany.
- Bezborodov A (1978) On secondary metabolites: Their functions and biogenesis. Folia Microbiol 23: 509-510.
- Maplestone RA, Stone MJ, Williams DH (1992) The evolutionary role of secondary metabolites-a review. Gene 115: 151-157.
- Haas D, Keel C (2003) Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117-153.
- Beneduzi A, Ambrosini A, Passaglia LM (2012) Plant growth promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet MolBiol 35: 1044-1051.
- Leisinger T, Margraff R (1979) Secondary metabolites of the fluorescent pseudomonds. Microbiol Rev 43: 422-442.
- Wang Y, Newman DK (2008) Redox reactions of phenazine antibiotics with ferric (hydr) oxides and molecular oxygen. Environ SciTechnol42: 2380-2386.
- Fitzpatrick DA (2009) Lines of evidence for horizontal gene transfer of a phenazine producing operon into multiple bacterial species. J MolEvol 68: 171-185.
- Mavrodi DV, Blankenfeldt W, Thomashow LS (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44: 417-445.
- Giddens SR, Feng Y, Mahanty HK (2002) Characterization of a novel phenazine antibiotic gene cluster in Erwiniaherbicola Eh1087. MolMicrobiol 45: 769-783.
- Smirnov V, Kiprianova E (1990) Bacteria of Pseudomonas genus. NaukovaDumka, Kiev, Ukraine.
- Thomashow LS, Weller DM (1988) Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomycesgraminis var. tritici. J Bacteriol 170: 3499-3508.
- Thomashow LS, Weller DM, Bonsall RF, Pierson LS (1990) Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol 56: 908-912.
- Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak YS, et al. (2011) Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl Environ Microbiol 1-11.
- Wang Y, Kern SE, Newman DK (2010) Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol 192: 365-369.
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, et al. (2011) Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193: 3606-3617.
- Singh IP, Sidana J, Bharate SB, Foley WJ (2010) Phloroglucinol compounds of natural origin: Synthetic aspects. Nat Prod Rep 27: 393-416.
- Achkar J, Xian MF, Zhao H, Frost J (2005) Biosynthesis of phloroglucinol. J Am ChemSoc 127: 5332-5333.
- Murata M, Yamakoshi Y, Homma S, Arai K, Nakamura Y (1992) Macrocarpals, antibacterial compounds from eucalyptus, inhibit aldose reductase. BiosciBiotechnolBiochem 56: 2062-2063.
- Iavicoli A,Boutet E, Buchala A, Métraux JP (2003) Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16: 851-858.
- Brazelton JN, Pfeufer EE, Sweat TA, McSpadden Gardener BB, Coenen C (2008) 2,4-diacetylphloroglucinol alters plant root development. Mol Plant Microbe Interact 21: 1349-1358.
- Fillinger S, Ajouz S, Nicot PC, Leroux P, Bardin M (2012) Functional and structural comparison of pyrrolnitrin-and iprodione-induced modifications in the class III histidine-kinase Bos1 of Botrytis cinerea. Plos One 7: 1-7.
- 108. Wong DT, Airall JM (1970) The mode of action of antifungal agents: Effect of pyrrolnitrin on mitochondrial electron transport. J Antibiot 23: 55-62.
- Wong DT, Airall JM (1970) The mode of action of antifungal agents: Effect of pyrrolnitrin on mitochondrial electron transport. J Antibiot 23: 55-62.
- Arima K, Imanaka H, Kousaka M, Fukuta A, Tamura G (1964) Pyrrolnitrin, a new antibiotic substance, produced by Pseudomonas. AgricBiolChem 28: 575-576.
- Hamill R, Elander R, Mabe J, Gorman M (1970) Metabolism of tryptophan by Pseudomonas aureofaciensIII. Production of substituted pyrrolnitrins from tryptophan analogues.ApplMicrobiol 19: 721-725.
- Howell C, Stipanovic R (1979) Control of Rhizoctoniasolani on cotton seedlings with Pseudomonasfluorescens and with an antibiotic produced by the bacterium. Phytopathology 69: 480-482.
- Nowak-Thompson B, Hammer PE, Hill DS, Stafford J, Torkewitz N, et al. (2003) 2, 5-Dialkylresorcinol biosynthesis in Pseudomonas aurantiaca: Novel head-to-head condensation of two fatty acid-derived precursors. J Bacteriol 185: 860-869.
Citation: Premachandra D, Hudek L, Brau L (2016) Bacterial Modes of Action for Enhancing of Plant Growth. J Biotechnol Biomater 6:236. Doi: 10.4172/2155-952X.1000236
Copyright: © 2016 Premachandra D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 22112
- [From(publication date): 9-2016 - Apr 25, 2024]
- Breakdown by view type
- HTML page views: 20471
- PDF downloads: 1641