Dr. Yunjo Soh
E-mail: ysoh@jbnu.ac.kr
Tel.: +82 63 270 4038, Fax: +82 63 270 4037
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 4
Page No: 738-747
Dr. Yunjo Soh
E-mail: ysoh@jbnu.ac.kr
Tel.: +82 63 270 4038, Fax: +82 63 270 4037
Mahesh Sapkotaa, Young Ran Parka, Se-Woong Kima, Yunjo Soha,*
a Department of Dental Pharmacology, School of Dentistry, Chonbuk National University, Jeon-Ju,561-756, Republic of Korea
Restituto Tocmo(tocmo@wisc.edu)
Jumin Hou(houjumin0511@126.com)
Xinwei Jiang(haiyuanjxw@126.com)
Yunjo Soh, Mahesh Sapkota, Young Ran Park, Se-Woong Kim Ran, Ras-related nuclear protein, regulates chondrogenic differentiation of ATDC5 cells(2019)SDRP Journal of Food Science & Technology 4(4)
Ras-related nuclear protein (Ran) is involved in cell regulation, nuclear-cytoplasmic transportation, and transformation and has been associated with the expression, occurrence, and progression of breast cancer, lung cancer, and colon cancer. We examined whether Ran is involved in chondrogenic differentiation in murine prechodrogenic ATDC5 cells, which can be differentiated into chondrocytes. In present study, we found that the expression of most chondrogenic marker genes including Aggrecan, BSP, Runx-2, Collagen I, Collagen II, and Collagen X, gradually increased for 14 days. Subsequently, we determined the mRNA and protein expression of Ran by using RT-PCR and western blot analysis. During chondrogenic differentiation, mRNA and protein expressions of Ran were significantly upregulated in ATDC5 cells. Next, Ran-siRNA transfection into ATDC5 cells was performed to knock down the expression of Ran during chondrogenic differentiation. We found that knockdown of Ran significantly suppressed size and number of cartilage nodules as well as the activity of alkaline phosphatase without cell cytotoxicity. Moreover, Ran knockdown significantly decreased not only mRNA expression of chondrogenic markers such as Aggrecan, BSP, Sox-9, Smad4, collagen I, II, X and OCN but also protein expression of ALP, OSX, BMP2, and Collagen 1A during chondrogenic differentiation. These results demonstrate that Ran could play a pivotal role in the transition of chondrocytes into hypertrophic chondrocytes and a mineralized phenotype.
Key words: ATDC5 cells, Ran, chondrogenesis, ALP, BMP2, collagen II.
Chondrogenic differentiation is an important process in ossification that occurs during development, growth, and renovation of the skeleton. Ossification involves several stages: resting, proliferation, maturation/hypertrophy, calcification, and ossification. During the early stages, chondrocytes proliferate rapidly and synthesize extracellular matrix (ECM). Sox9 induces chondrocyte differentiation as a master transcription factor [1, 2]. Differentiated chondrocytes produce cartilage-specific matrix proteins such as collagen type II and Aggrecan, and then undergo hypertrophy and terminal differentiation due to secretion of collagen type X [3, 4]. BMP2 induces both early-phase and late-phase chondrogenic differentiation [2, 5]. BMP2 also interacts with Runx2 at the post-transcriptional level [6]. Runx2 plays a central role in chondrocyte maturation and osteogenesis [7].
Small G-proteins are monomeric G-proteins with a molecular mass ranging from 20-40 kDa. Two genes, Ha-Ras and Ki-Ras are oncogenes of the sarcoma virus that were discovered almost four decades ago [8]. To date, more than 100 G proteins have been recognized in humans and eukaryotes, and they are included in a superfamily comprising five families: Rab, Ras, Rho, Sar1/Arf, and Ran families [9]. Small G proteins have major functional and structural roles in different cells. Rab proteins are present in most eukaryotic cells and are the largest G protein family. More than 50 Rab proteins are found in mammals [10, 11]. Among them, Rab23 plays a crucial role in chondrogenesis by suppressing the expression of chondrogenic markers such as collagen type II, Aggrecan, and Sox9 [12]. There are three different Ras proteins capable of transforming mammalian cells by point mutation activation [13, 14]. Ras proteins can have carcinogenic activity as they control various extracellular pathways for the growth of cells, differentiation, and apoptosis [15, 16]. Rho small G proteins, containing RhoA, Rac1, and Cdc42, upregulate expression of proteins involved in the actin cytoskeleton as well as those involved in cell cycle progression [17]. Rac1/Cdc42 and RhoA promote mesenchymal cell differentiation and control the expression of chondrocyte-specific genes [18-20]. RhoA/ROCK suppresses chondrogenic activity in ATDC5 cells and mesenchymal cells [21].
Ras-related nuclear protein, Ran, regulates nucleocytoplasmic transport and microtubule organization during G1, S, G2 and M phases of the cell cycle [22]. Ran is also involved in nuclear transport, condensation of chromatin, formation of mitotic spindles, and formation of the post-mitotic nuclear assembly [23]. This protein has also been shown to play a pivotal role in cancer occurrence, development, and progression [24, 25]. Moreover, Ran was shown to have inhibitory effects in the treatment of cancer cells [26]. Inhibition of Ran can decrease cytoplasmic and nuclear accumulation by inducing cell senescence in embryonic fibroblasts [27]. However, the role of Ran in chondrogenesis has not been reported. In this study, we investigated the role of Ran in ATDC5 cells during chondrogenic differentiation by knockdown experiments. We found that knockdown of Ran suppressed the expression of chondrogenic marker genes, chondrogenic signaling molecules, ALP activity, and alcian blue staining in-vitro. These results suggest that Ran plays a crucial role in chondrocyte hypertrophy and mineralization.
2.1. Reagents
Ran siRNA and control siRNA were purchased from Ambion (Foster City, CA USA). Anti-Ran, anti-Col 1, anti-ALP, anti-OSX, and anti-BMP-2 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA USA). Anti-Runx2 was brought from Bioworld Technology, Inc. (Bioworld, MN USA). Fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Gibco (Grand Island, NY, USA). β-actin, 0.25% trypsin-EDTA, and other chemicals were purchased from Sigma (St. Louis, MO, USA). Other reagents were obtained as mentioned previously [28, 29].
2.2. Cell Culture and chondrogenic differentiation
ATDC5 cells were obtained from Riken Bio Research Center Cell Bank (Ibaragi, Japan). ATDC5 cells were cultured in Dulbecco’s modified Eagle’s medium and Ham’s F-12 Medium (DMEM/F-12) (1:1 ratio) supplemented with 5% FBS, 100 U/ml penicillin, streptomycin at 37°C in the presence of 5% CO2 and 95% air. During chondrogenic differentiation, ATDC5 cells were cultured in 35-mm plates at a density of 3.5x105 cells per plate in differentiation medium containing α-MEM with 10 µg/ml bovine insulin, 10 µg/ml human transferrin, 3x10-8 sodium selenite, and 50 ng/ml ascorbic acid at 37°C in the presence of 5% CO2 and 95% air for 14 days. Culture medium was changed every 48 h.
2.3. MTT Assay
Cell viability was examined by MTT assay. ATDC5 cells were seeded in 48-well plates at a density of 4 x105 cells/well. The next day, cells were transfected with scrambled siRNA or Ran siRNA for 48 h. After 48 h, cells were incubated with MTT for 2 h at 37°C. Then, cells were washed with 1 x PBS and 200 µl DMSO was added. Cell viability was determined by measuring the absorbance at 540 nm with a BioTek (Power wave HT, USA) microplate reader.
2.4. Alcian blue staining
ATDC5 cells were seeded in 48-well plates at a density of 3x 104 cells/well. The next day, cells were transfected with control siRNA or Ran siRNA for 48 hr. Then, the medium was replaced with differentiation medium and cells were incubated for 12 days. After 12 days, the ATDC5 cells were rinsed in 1x PBS three times, fixed with 4% paraformaldehyde for 15 min at -20°C, and stained with 1% alcian blue 8GS (Sigma, St. Louis, MO, USA) overnight at room temperature. Next, 3% acetic acid was used to wash ATDC5 cells three times. Images were taken with a digital camera. For quantification, stained cells were extracted with 10% acetic acid overnight at room temperature. Absorbance was measured at 650 nm using a BioTek plate reader (Power wave HT).
2.5. ALP activity
For ALP activity, ATDC5 cells were lysed with lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA) and collected in micro-centrifuge tubes. After incubation at 4 °C for 10 min, cells were centrifuged at 15,000 rpm for 15 min and supernatant was collected to protein concentration. ALP activity was measured using para-nitrophenyl-phosphate substrate (N7653, Sigma, USA) as a substrate. Briefly, equal sample was added with pNPP (1 mg/ml) in 1M diethanolamine buffer containing 0.5 mM MgCl2 (pH 9.8) at 37 °C for 20 min. The reaction was blocked by adding 200 µl of 2 M NaOH to the reaction mixture. Absorbance at 405 nm was measured and the protein concentration of cell lysates was determined using the Bradford assay at 595 nm using a BioTek plate reader (Power wave HT). ALP activity was normalized according to the total protein concentration. Experiments were performed in triplicate and the significance level was set at P<0.05.
2.6. Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cultured cells using TRIzol (Invitrogen, Life Technology), and cDNA was amplified using Super Script II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol and stored at -80°C. Primer sequences and annealing temperatures used in this study are listed in Table 1. Initial denaturation was performed for 1 min at 95°C, followed by 30 cycles of 94°C for 30 sec, annealing at various temperatures for 1 min, and extension at 72°C for 2 min. Taq polymerase was used (Promega, Madison, WI, USA). Reaction products were mixed with ethidium bromide and a 1% agarose gel was used to separate the reaction products. UV light (Gel-Doc, Bio-Rad, Hercules, CA, USA) was used to observe DNA bands. Phosphoimager and Quality One software (Version 4.3.1) (Bio-Rad) were used to analyze DNA band intensity.
Table 1.Primer sequences and conditions for RT-PCR
|
Target genes (Accession number) |
Primer (Forward, Reverse) |
Anneling Tm(°C) |
PCR cycles |
|
Aggrecan (NM_007424) |
5′-catgagaggccaatggaacg-3′ 5′-gaatcacctgcacagacccaa-3′ |
55 |
27 |
|
Collagen 1 (NM_007742) |
5′-ttctcctggtaaagatggtgc-3′ 5′-tgttaaaggtgatgctggtcc-3′ |
50 |
35 |
|
Collagen II (NM_031163) |
5′-ctgtaagaacagcatcgcctacctg-3′ 5′-caggaatttggtgtggacataggg-3′ |
60 |
27 |
|
Collagen X (NM_009925) |
5′-cgtctcgcttttactgtca-3′ 5′-ctcacagaaaatgaccaggt-3′ |
48 |
35 |
|
Runx-2 (NM_009820) |
5′-actttctccaggaagactgc-3′ 5′-acagcaacagcaacaacagc-3′ |
50 |
35 |
|
ALP (NM_007431) |
5′-ccatggtagattacgctcaca-3′ 5′-atggaggattccagatacagg-3′ |
56 |
30 |
|
OCN (NM_001032298) |
5′- cagcttggtgcacacctagc-3′ 5′- ggagcagtgtcacgttaacct-3′ |
58 |
30 |
|
BSP (NM_008318) |
5′-gagccaggactgccgaaaggaa-3′ 5′-gcagcagcggaggagaaacgg-3′ |
60 |
27 |
|
Smad4 (AY493561) |
5′-atctatgcccgtctgtggag-3′ 5′-aacattcctgtggcttccac-3′ |
50 |
30 |
|
SOX-9 (NM_011448) |
5′-cacaagaaagaccaccccgatac-3′ 5′-ggcaaagttgatctgaagcgaga-3′ |
60 |
32 |
|
β-actin (NM_007393) |
5′-ttctacaatgagctgcgtgt-3′ 5′-ctcatagctcttctccaggg-3′ |
50 |
26 |
2.7. Immunoblot analysis
ATDC5 cells were harvested and washed with 1x PBS three times, after which total protein was extracted using lysis buffer [20 mM Tris–HCl (pH 7.5), 137 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM Na3VO4, 1 mM phenyl methyl sulfonyl fluoride (PMSF), and 1 X protease inhibitor cocktail]. Protein assay dye (Bio-Rad) was used to measure the protein concentration using BSA as a standard. Same amounts of total protein were separated by 10% SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and blocked with 5% skim milk in Tris-buffer saline (TBS) with 0.25% Tween-20 (TTBS) for 1 h at room temperature. This was followed by an overnight incubation at 4°C with the following primary antibodies: anti-Ran (Santa Cruz Biotechnology, Santa Cruz, CA), anti-ALP (Santa Cruz Biotechnology), anti-Osx (Santa Cruz Biotechnology), anti-Runx2 (Santa Cruz Biotechnology), or anti-col I (Santa Cruz Biotechnology) at (1:1500) and anti-β-actin (1:4000; Santa Cruz Biotechnology), followed by anti-rabbit, anti-mouse, or anti-goat (1:3000; Santa Cruz Biotechnology) for 1 h at room temperature. After washing with TBST, Clarity Western ECL Substrate (Bio-Rad) and medical X-ray film (Fuji Photo Film, Tokyo, Japan) were used to detect immune-reactive bands. Quantitative analysis was performed using Image J software and the optical intensities of individual band were normalized to the corresponding values of β- actin.
2.8. siRNA transfection
ATDC5 cells (4 x 105 cells) were seeded in 35-mm plates and cultured in α-MEM with 10% FBS without antibiotics overnight. ATDC5 cells were transfected with 100 nM siRNA using Lipofectamine RNAiMAX and scrambled siRNA (Thermo Fisher Science Life Science) was used as a control. ATDC5 cells were transfected for 48 h, which did not affect cellular and molecular function. After 48 h, cells were cultured in differentiation medium to induce chondrogenic differentiation before subsequent experiments.
2.9 Statistical analysis
Results are expressed as mean values ± standard deviations (SD). One-way ANOVAs with Tukey’s multiple comparison tests were performed using Graphpad prism software (Graphpad Software Inc., La Jolla CA, USA). All experiments were repeated three times and *P<0.05 and **P<0.01 were considered to indicate statistically significant results.
3.1 ATDC5 cells differentiate into chondrogenic cells
ATDC5 cells were seeded in differentiation medium for 0, 7, and 14 days to induce chondrogenic differentiation, and alcian blue staining was performed. Nodule formation was detected during chondrogenic differentiation. (Fig. 1A). To explore the expression level of chondrogenic marker genes such as ALP, Aggrecan, BSP, Sox-9, Runx-2, Smad 4, collagen I, collagen II, and collagen X, RT-PCR was performed at 0, 7, and 14 days (Fig. 1B). mRNA expression levels of Aggrecan, BSP, Runx-2, Collagen I, Collagen II, and Collagen X increased during chondrogenic differentiation and peaked at 14 days, while those of ALP, Sox-9, and Smad4 remained unchanged. (Fig. 1C-K). These results indicate that chondrogenic differentiation was efficiently induced in ATDC5 cells.
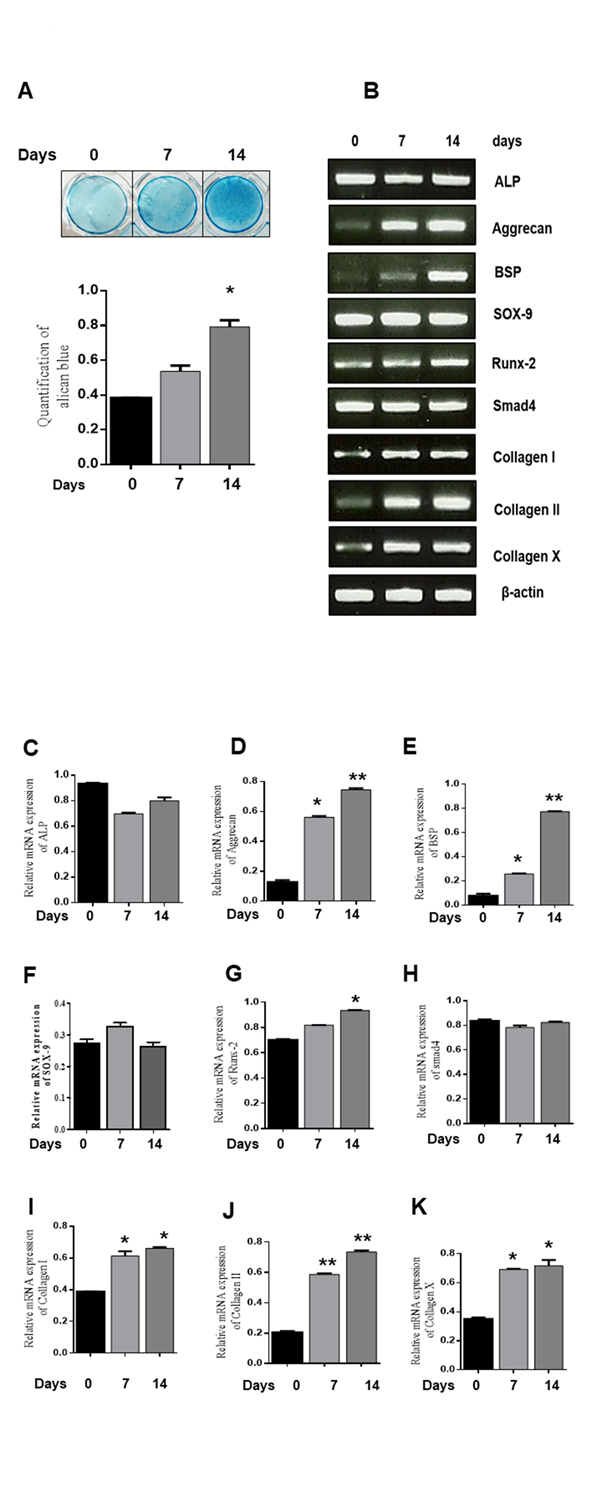
Figure 1. Chondrogenic differentiation of ATDC5 cells. ATDC5 cells were seeded in 35-mm plates and cultured in α-MEM supplemented with 10 µg/ml bovine insulin, 10 µg/ml human transferrin, 3x10-8 sodium selenite (ITS), and 50 ng/ml ascorbic acid. (A) Alcian blue staining was observed and quantified. *P < 0.05 indicates a significant difference compared to the day 0. (B-K) mRNA expression of chondrogenic marker genes at 0, 7, and 14 days was determined by RT-PCR. Relative mRNA expression levels of various genes. β-actin was used as an internal control. Values are expressed as means ± S.D., n=3. *P<0.05, **P<0.01 indicates a significant difference compared to the control.
3.2 Ran mRNA and protein expression increase during chondrogenic differentiation of ATDC5 cells
We examined whether Ran is involved in chondrogenic differentiation by analyses of mRNA and protein expression levels using RT-PCR and western blot. We found that Ran mRNA and protein expression markedly increased during chondrogenic differentiation compared with control (Fig. 2A, B). To determine the expression of Ran on chondrogenic differentiation in detail, we transfected ATDC5 cells with Ran siRNA for 0, 7, and 14 days. As shown in Fig. 2C and 2D, knockdown of Ran decreased Ran mRNA and protein expression during chondrogenic differentiation. These results support that Ran induces chondrogenic differentiation in ATDC5 cells.
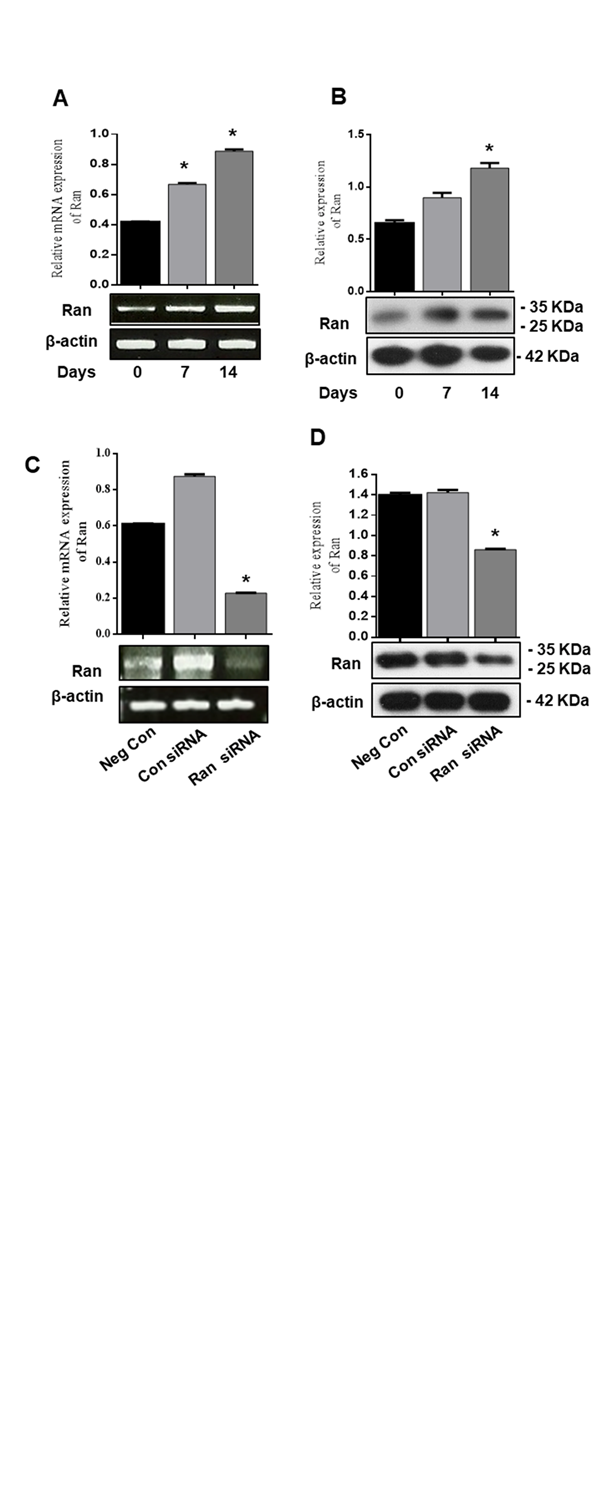
Figure 2. Ran mRNA and protein expression increased during chondrogenic differentiation of ATDC5 cells. ATDC5 cells were seeded in 35-mm plates and cultured in α-MEM supplemented with 10 µg/ml bovine insulin, 10 µg/ml human transferrin, 3x10-8 sodium selenite (ITS), and 50 ng/ml ascorbic acid. (A) mRNA expression of Ran at 0, 7, and 14 days was determined by RT-PCR. Relative mRNA expression level of Ran. β-actin was used as the internal control. (B) Protein expression of chondrogenic markers at 0, 7, and 14 days was determined by western blot. Relative density of Ran. β-actin was used as an internal control. ATDC5 cells were transfected with control siRNA or Ran siRNA for 48 h. (C) mRNA expression and (D) protein expression were determined by RT-PCR and western blot analysis. Values are expressed as means ± S.D., n=3. *P<0.05 indicates a significant difference compared to the control.
3.3 Ran knockdown suppresses alcian blue Staining and ALP activity
To examine whether Ran is involved in chondrogenic differentiation, ATDC5 cells were transfected with 100 nM scrambled siRNA or 100 nM Ran siRNA for 48 h and then incubated in differentiation medium for 12 days. The nodule formation was observed during chondrogenic differentiation by using alcian blue staining (Fig. 3A). Alcian blue staining intensity was also markedly decreased in Ran knockdown cells compared to those transfected with scrambled siRNA (Fig. 3B). Expression of ALP is an indication of mineralization [30]. To determine ALP activity, ATDC5 cells were transfected with 100 nM scrambled siRNA or 100 nM Ran siRNA for 48 h followed by incubation in differentiation medium for 12 days. ALP activity was significantly inhibited in cells transfected with Ran siRNA (Fig. 3C). Moreover, control siRNA or Ran siRNA (25, 50, 100 nM) were transfected to ATDC5 cells for 48 h and knockdown of Ran at indicated concentrations showed no cytotoxic effects on ATDC5 cells compared with negative control siRNA (Fig. 3D). Therefore, 100 nM Ran was selected for further experiments.
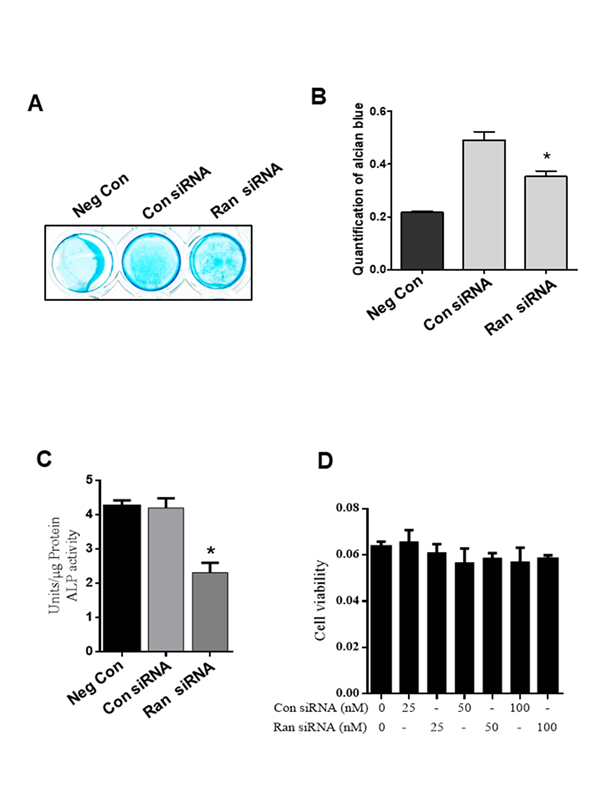
Figure 3. Effect of Ran knockdown on proteoglycan production in ATDC5 cells. (A) Alcian blue staining was observed and (B) quantified. (C) ALP activity was measured in cells transfected with control siRNA. (D) ATDC5 cells were transfected with various concentration of control siRNA and Ran siRNA (25, 50, 100 nM) for 48 h and MTT assay was performed. Values are expressed as means ± S.D., n=3. *P<0.05 indicates a significant difference compared to control siRNA.
3.4 Ran knockdown inhibits mRNA expression during chondrogenic differentiation in ATDC5 cells
To identify whether Ran functions in chondrogenic differentiation, ATDC5 cells were transfected with 100 nM scrambled siRNA or 100 nM Ran siRNA for 48 h, and then the medium was replaced with differentiation medium and cells were incubated for 12 days. Knockdown of Ran markedly inhibited the expression of Aggrecan, BSP, SOX-9, Collagen I, Collagen II, Collagen X, and OCN, but not Runx2 (Fig. 4B-J). These results suggest that Ran could play an important role in chondrogenesis.
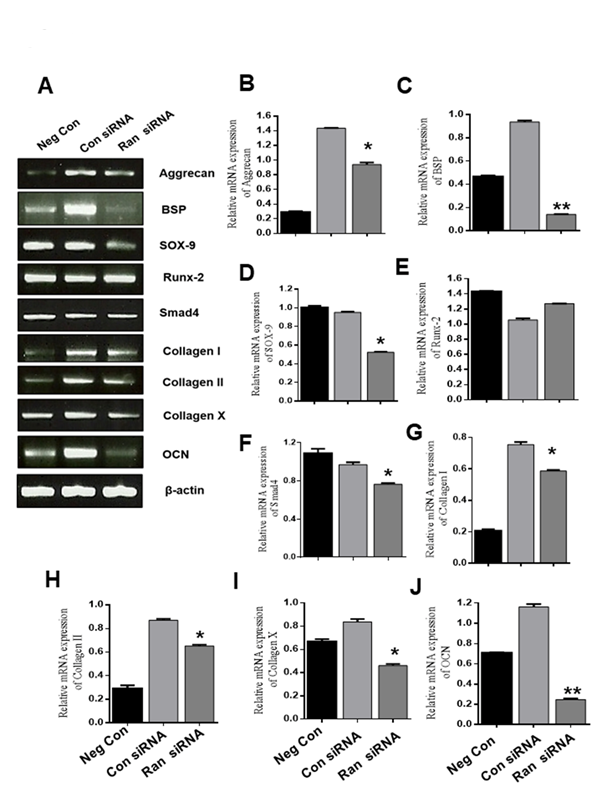
Figure 4. Effect of Ran knockdown on mRNA levels during chondrocyte differentiation. (A) mRNA expression of chondrogenic marker genes was assessed by RT-PCR. β-actin was used as the internal control. (B-J) Quantification of various chondrogenic marker genes. Values are expressed as means ± S.D., n=3. *P < 0.05, **P <0.01 indicates a significant difference compared to the control.
3.5 Ran knockdown represses protein expression during chondrogenic differentiation of ATDC5 cells
We next investigated the protein expression of chondrogenic differentiation markers (Fig. 5A). ATDC5 cells were transfected with 100 nM scrambled siRNA or 100 nM Ran siRNA for 48 h and then transferred to differentiation medium for 12 days. We found that Ran knockdown suppressed the expression of ALP, OSX, and Col I, but not Runx2 (Fig. 5A-E). These results confirm that Ran plays a crucial role in chondrogenic protein expression.
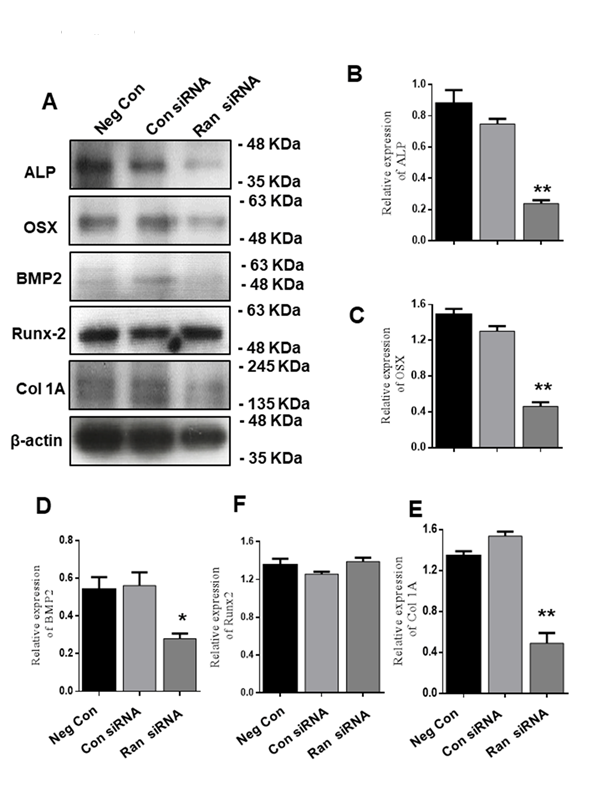
Figure 5. Effect of Ran knockdown on protein levels of chondrocyte differentiation markers during chondrocyte differentiation. (A) Protein expression of chondrogenic marker genes was evaluated by western blot. β-actin was used as an internal control. (B-E). Quantification of various proteins. Values are expressed as means ± S.D., n=3. *P<0.05, **P<0.01 indicate a significant difference compared to the control.
Chondrogenesis is an active biological process that involves tissue patterning and development of skeletal and endochondral ossification [3]. Condensation of multipotent mesenchymal cells is the primary stage of chondrogenesis, after which cells proliferate and differentiate into chondrocytes [31]. Proliferative chondrocytes produce chondrogenic matrices together with aggrecan and collagen and differentiate into hypertrophic chondrocytes [3]. Extracellular matrix (ECM) is composed of proteoglycans, and interactions between the ECM and chondrocytes help regulate cell growth, differentiation, and morphogenesis [19, 32]. We hypothesized that Ran may be involved in chondrocyte differentiation, pre-hypertrophy, and hypertrophy.
Insulin-transferrin-selenium added to a growth medium induces chondrogenesis in various cell lines [33, 34]. In this study, we investigated the expression levels of various chondrogenic marker genes at different time points and found a significant increase in chondrogenic marker gene expression at 14 days after incubation of ATDC5 cells in differentiation medium. Ran mRNA and protein levels were also markedly increased of Ran at 7 and 14 days compared to control cells not incubated in differentiation medium.
Knockdown of Ran significantly suppressed the mRNA expression of most chondrogenic differentiation marker. The relationships among aggrecan, Ran, collagen II, and collagen X in chondrocytes remain unclear. Various studies have shown that aggrecan and collagen II are important proteoglycan elements in the ECM of articular cartilage that are required for proliferation and synthesis [4, 35]. Collagen X was shown to stop chondrocyte proliferation and promote chondrocyte enlargement [36]. Sox9 was found to be expressed in premature chondrocytes during chondrocyte differentiation [37]. While smad4 was expressed during proliferation and hypertrophic chondrocyte cartilage development [38]. BMP-2 is involved in both early and late phases of chondrogenic differentiation in ATDC5 cells [5, 39]. Osteocalcin plays a pivotal role in chondrocyte cell differentiation and is also involved in cartilage mineralization [40]. Runx2 is a transcriptional factor that plays an important role in promoting prehypertrophic cells to become hypertrophic. Runx2 is also involved in chondrocyte maturation before mineralization [7, 37]. Various studies have suggested that osterix may play a regulatory role in cartilage formation, and that a deficiency of osterix can affect bone formation [41, 42]. In the current study, knockdown of Ran suppressed the mRNA expression of Ran, aggrecan, BSP, SOX-9, Smad4, collagen I, collagen II, collagen X, and OCN but not the expression of Runx2. Moreover, Ran knockdown suppressed the protein expression of Ran, ALP, OSX, BMP2, and collagen I but not the expression of Runx-2. Likewise, the intensity of alcian blue staining was markedly reduced in Ran knockdown ATDC5 cells. ALP is a marker of prehypertrophic chondrocytes [43, 44], which are associated with differentiation, chondrocyte accumulation in the ECM, and mineralization [45, 46]. Ran knockdown inhibited ALP activity on day 12, which suggested that Ran participates in late stage chondrogenic differentiation during mineralization. Further studies are needed to arrive at a definitive conclusion whether overexpression of Ran promote the differentiation of chondrocytes in ATDC5 cells, or in vivo assay.
In summary, our results indicate that Ran regulates pre-hypertrophy, hypertrophy, and mineralization in ATDC5 cells with a chondrogenic phenotype. Therefore, Ran may be a possible therapeutic target in chondrogenic disorders.
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF- 2017R1A2B4011988), Research Base Construction Fund Support Program funded by Chonbuk National University in 2018
Akiyama H, Chaboissier MC, Martin JF, Schedl A. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813-28. PMid:12414734
View Article PubMed/NCBIAkiyama H, Shukunami C, Nakamura T, Hiraki Y. Differential expressions of BMP family genes during chondrogenic differentiation of mouse ATDC5 cells. Cell Struct Funct. 2000;25:195-204. PMid:10984103
View Article PubMed/NCBIKronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332-6. PMid:12748651
View Article PubMed/NCBIO'Keefe RJ, Puzas JE, Loveys L, Hicks DG. Analysis of type II and type X collagen synthesis in cultured growth plate chondrocytes by in situ hybridization: rapid induction of type X collagen in culture. J Bone Miner Res. 1994;911:1713-22. PMid:7863822
View Article PubMed/NCBIShukunami C, Akiyama H, Nakamura T, Hiraki Y. Requirement of autocrine signaling by bone morphogenetic protein-4 for chondrogenic differentiation of ATDC5 cells. FEBS Lett. 2000;469:83-7. 01251-5
View ArticleShu B, Zhang M, Xie R, Wang M. BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J Cell Sci. 2011;124:3428-40. PMid:21984813
View Article PubMed/NCBIYoshida CA, Yamamoto H, Fujita T, Furuichi T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952-63. PMid:15107406
View Article PubMed/NCBIChien UH, Lai M, Shih TY, Verma IM. Heteroduplex analysis of the sequence relationships between the genomes of Kirsten and Harvey sarcoma viruses, their respective parental murine leukemia viruses, and the rat endogenous 30S RNA. J Virol. 1979;31:752-60.
Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125-32. PMid:2122258
View Article PubMed/NCBINovick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496-504. 80025-7
View ArticleNuoffer C, Balch WE. GTPases: multifunctional molecular switches regulating vesicular traffic. Annu Rev Biochem. 1994;63:949-90. PMid:7979258
View Article PubMed/NCBIYang L, Clinton JM, Blackburn ML, Zhang Q. Rab23 regulates differentiation of ATDC5 chondroprogenitor cells. J Biol Chem. 2008;283:10649-57. PMid:18218620
View Article PubMed/NCBIBrown R, Marshall CJ, Pennie SG, Hall A. Mechanism of activation of an N-ras gene in the human fibrosarcoma cell line HT1080. EMBO J. 1984;3:1321-6 PMid:6086315
View Article PubMed/NCBIFeramisco JR, Gross M, Kamata T, Rosenberg M. Microinjection of the oncogene form of the human H-ras (T-24) protein results in rapid proliferation of quiescent cells. Cell. 1984;38:109-17. 90531-2
View ArticleRodenhuis S, Slebos RJ, Boot AJ, Evers SG. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988;48:5738-41.
Sriskanthadevan-Pirahas S, Deshpande R, Lee B, Grewal SS. Ras/ERK-signalling promotes tRNA synthesis and growth via the RNA polymerase III repressor Maf1 in Drosophila. PLoS Genet. 2018;14:e1007202. PMid:29401457
View Article PubMed/NCBIEtienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629-35. PMid:12478284
View Article PubMed/NCBIKim H, Sonn JK. Rac1 promotes chondrogenesis by regulating STAT3 signaling pathway. Cell Biol Int. 2016;40:976-83. PMid:27306109
View Article PubMed/NCBIWoods A, Wang G, Beier F. Regulation of chondrocyte differentiation by the actin cytoskeleton and adhesive interactions. J Cell Physiol. 2007;213:1-8. PMid:17492773
View Article PubMed/NCBIWoods A, Wang G, Dupuis H, Shao Z. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J Biol Chem. 2007;282:23500-8. PMid:17573353
View Article PubMed/NCBIWoods A, Wang G, Beier F. RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J Biol Chem. 2005;280:11626-34. PMid:15665004
View Article PubMed/NCBIGarcia-Ranea JA, Valencia A. Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 1998;434:219-25 00967-3
View ArticleDasso M. The Ran GTPase: theme and variations. Curr Biol. 2002;12:R502-8. 00970-3
View ArticleAbe H, Kamai T, Shirataki H, Oyama T. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer. 2008;122:2391-7. PMid:18241036
View Article PubMed/NCBIXia F, Canovas PM, Guadagno TM, Altieri DC. Mol Cell Biol. 2008;28:5299-311. PMid:18591255
View Article PubMed/NCBIYuen HF, Chan KK, Grills C, Murray JT. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res 2012;18:380-91. PMid:22090358
View Article PubMed/NCBINagai M, Yoneda Y. Downregulation of the small GTPase ras-related nuclear protein accelerates cellular ageing. Biochi Biophys Acta. 2013;1830:2813-9. PMid:23160023
View Article PubMed/NCBILi L, Sapkota M, Gao M, Choi H. Macrolactin F inhibits RANKL-mediated osteoclastogenesis by suppressing Akt, MAPK and NFATc1 pathways and promotes osteoblastogenesis through a BMP-2/smad/Akt/Runx2 signaling pathway. Eur J Pharmacol. 2017;815:202-9. PMid:28919027
View Article PubMed/NCBISapkota M, Li L, Choi H, Gerwick WH. Bromo-honaucin A inhibits osteoclastogenic differentiation in RAW 264.7 cells via Akt and ERK signaling pathways. Eur J Pharmacol. 2015;769:100-9. PMid:26550952
View Article PubMed/NCBIOrimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch. 2010;77:4-12. PMid:20154452
View Article PubMed/NCBIPass C, MacRae VE, Ahmed SF, Farquharson C. Inflammatory cytokines and the GH/IGF-I axis: novel actions on bone growth. Cell Biochem Funct. 2009;27:119-27. PMid:19330796
View Article PubMed/NCBIRozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Deve Biol. 2010;341:126-40. PMid:19854168
View Article PubMed/NCBIChua KH, Aminuddin BS, Fuzina NH, Ruszymah BH. Insulin-transferrin-selenium prevent human chondrocyte dedifferentiation and promote the formation of high quality tissue engineered human hyaline cartilage. Eur Cells Mater. 2005;9:58-67
View ArticleLiu X, Liu J, Kang N, Yan L. Role of insulin-transferrin-selenium in auricular chondrocyte proliferation and engineered cartilage formation in vitro. Int J Mol Sci. 2014;15:1525-37. PMid:24451136
View Article PubMed/NCBIKravis D, Upholt WB. Quantitation of type II procollagen mRNA levels during chick limb cartilage differentiation. Dev Biol. 1985;108:164-72. 90018-1
View ArticleKozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817-31. PMid:25715393
View Article PubMed/NCBIZhong L, Huang X, Karperien M, Post JN. The Regulatory Role of Signaling Crosstalk in Hypertrophy of MSCs and Human Articular Chondrocytes. Int J Mol Sci. 2015;16:19225-47. PMid:26287176
View Article PubMed/NCBIXiao D, Wang R, Hu J, Quan H. Spatial and temporal expression of Smad signaling members during the development of mandibular condylar cartilage. Exp Ther Med. 2017;14:4967-71. PMid:29201201
View Article PubMed/NCBIWahl M, Shukunami C, Heinzmann U, Hamajima K. Transcriptome analysis of early chondrogenesis in ATDC5 cells induced by bone morphogenetic protein 4. Genomics. 2004;83:45-58. 00201-5
View ArticleHan Y, Xu G, Zhang J, Yan M. Leptin induces osteocalcin expression in ATDC5 cells through activation of the MAPK-ERK1/2 signaling pathway. Oncotarget. 2016;7:64021-9. PMid:27564111
View Article PubMed/NCBINakashima K, Zhou X, Kunkel G, Zhang Z. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17-29. 00622-5
View ArticlePark SY, Kim JE. Differential gene expression by Osterix knockdown in mouse chondrogenic ATDC5 cells. Gene. 2013;518:368-75. PMid:23337593
View Article PubMed/NCBIChikuda H, Kugimiya F, Hoshi K, Ikeda T. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418-29. PMid:15466490
View Article PubMed/NCBINakatani S, Mano H, Sampei C, Shimizu J. Chondroprotective effect of the bioactive peptide prolyl-hydroxyproline in mouse articular cartilage in vitro and in vivo. Osteoarthritis Cartilage. 2009;17:1620-7. PMid:19615963
View Article PubMed/NCBIMackie EJ, Tatarczuch L, Mirams M. The skeleton: a multi-functional complex organ: the growth plate chondrocyte and endochondral ossification. J Endocrinol. 2011;211:109-21. PMid:21642379
View Article PubMed/NCBIShukunami C, Ohta Y, Sakuda M, Hiraki Y. Sequential progression of the differentiation program by bone morphogenetic protein-2 in chondrogenic cell line ATDC5. Exp Cell Res. 1998;241:1-11. PMid:9633508
View Article PubMed/NCBI