Matthias Stope,
Phone: +49-3834-86-80436; Fax: +49-3834-86-80435;
E-mail: matthias.stope@uni-greifswald.de
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 3 ISSUE: 1
Page No: 164-170
Matthias Stope,
Phone: +49-3834-86-80436; Fax: +49-3834-86-80435;
E-mail: matthias.stope@uni-greifswald.de
Hannes Ahrend1, Georg Daeschlein2, Elias Grove1, Madeleine Paditz3, Alexander Mustea3, Martin Burchardt1, Matthias B. Stope1*
1Department of Urology, University Medicine Greifswald, Greifswald, Germany
2Department of Dermatology, University Medicine Greifswald, Greifswald, Germany
3Department of Gynaecology and Obstetrics, University Medicine Greifswald, Greifswald, Germany
Farhadul Islam(farhad_bio83@ru.ac.bd)
Zhe-Feng Xiao(xiaozf@csu.edu.cn)
Peeyush K Lala(pklala@uwo.ca)
Hannes Ahrend et al., MicroRNA-20a-3p and microRNA-20a-5p exhibit anti-proliferative activities in a melanoma in vitro model(2019)SDRP Journal of Cellular and Molecular Physiology 3(1) p:164-170
MicroRNAs control numerous cancer-related signaling pathways and play pivotal role in cancer initiation and progression. Recent studies have indicated variable and cancer-specific expression patterns of microRNA-20a (miR-20a), which have been attended by varying and sometimes contrary tumor biological functions. This is the first study regarding to the characterization of miR-20a's functionality in melanoma cells.
miR-20a expression was examined by reverse transcriptase and quantitative polymerase chain reaction in an in vitro melanoma model containing HaCat keratinocytes and B16 melanoma cells. For cell growth analysis, miR-20a vectors were cloned and transfected into B16 cells. Cell growth kinetics were performed utilizing a Cell Counter and Analyzer Model TT (Roche Applied Science). The expression of both the 3p and the 5p strand processed from the miR-20a precursor was suppressed in melanoma cells B16 compared to the expression in non-malignant HaCat keratinocytes. Recombinant restoration of miR-20a levels in malignant B16 cells attenuated cellular growth. Our data suggest that miR-20a bears biological functions in melanoma cells and thus represents an anti-oncogenic factor which is suppressed during cancer progression.
Key Words: microRNA-20a, cancer, melanoma cells, keratinocytes, skin cancer model, tumor suppressor
Each microRNA controls the expression of up to hundreds of genes thereby modulating pivotal signaling cascades including cancer-related pathways. Compared to other malignancies, little is known about microRNA's dysregulation and biological efficacy in melanoma cells[1]. MicroRNA-21, for instance, represents a well-characterized tumor promotor and belongs to the few microRNAs which have been examined in skin cancer[2–5]. In case of microRNA-20a (miR-20a), little is known about its role in melanoma progression. In other cancer cells, recent studies have indicated highly variable and cancer-specific expression patterns of both the 3 prime (3p) as well as the 5 prime (5p) strand of the miR-20a precursor molecule (miR-20a-3p and miR-20a-5p). Certainly, the differentiation in the two miR-20a forms 3p and 5p is oftentimes being missed in molecular analysis thereby hindering analysis of the biological efficacy. In astrocytoma, miR-20a-5p expression is increased whereas the expression in glioblastoma cells is decreased[6,7]. In breast cancer, miR-20a is significantly downregulated compared with healthy tissue and the overexpression of miR-20a inhibits cellular proliferation[8]. Another study, however, demonstrated a 4-fold increase of miR-20a-5p in triple-negative breast cancer compared with luminal A breast invasive ductal carcinoma[9]. Furthermore, restoration of miR-20a leads to an attenuation of hepatocellular carcinoma growth but in contrast to the promotion of cervical cancer growth[10,11]. Beside proliferation control, miR-20a is also involved in chemoresistance mechanisms. The microRNA appears as an inhibitor of multi-drug resistance in osteosarcoma as well as an inducer of cisplatin resistance during gastric cancer therapy[12,13]. Interestingly, in case of hepatocellular and gastric cancer, miR-20a correlates with clinical parameter and thus is discussed as a promising biomarker[14,15].
The study presented here started to characterize miR-20a's proliferative properties applying an established melanoma model comprising non-malignant HaCat keratinocytes and B16 melanoma cells[16-19].
Cell Culture
Human keratinocytes HaCat (German Cancer Research Center (DKFZ), Heidelberg, Germany) and murine melanoma cells B16 (Cell Line Service, Eppelheim, Germany) were propagated in DMEM medium (PAN Biotech, Aidenbach, Germany) supplemented with 2 mM glutamine, 1% penicilline/streptomycine (Biochrom, Berlin, Germany) and 8% fetal bovine serum (Sigma-Aldrich, Deisenhofen, Germany) and DMEM medium (PAN Biotech) containing 4.5 g/l glucose, 2 mM glutamine, 1% penicilline/streptomycine (Biochrom), and 10% fetal bovine serum (Sigma-Aldrich), respectively. Both cell lines were cultivated in a humidified atmosphere at 37°C with 5% CO2.
Proliferation Assay
Cell growth was determined by cell counting (CASY Cell Counter and Analyzer Model TT, Roche Applied Science, Mannheim, Germany). Therefor, adherent cells were treated with trypsin/ethylenediaminetetraacetic acid (EDTA) and 1:100 diluted in CASYton (Roche Applied Science). Subsequently, 400 μl of the cell dilution was analyzed in triplicates. Measurement was performed applying a capillary of 150 μm in diameter and cell line-specific gate settings to discriminate between living cells, dead cells, and cellular debris: 6.6 μm/10.95 μm (HaCat), 7.8 μm/12.0 μm (B16).
RNA preparation and cDNA synthesis
Total RNA preparation was done using peqGOLDTrifast Reagent (Peqlab Biotechnology, Erlangen, Germany) according to the manufacturer’s instructions. RNA concentration was determined utilizing a Nanodrop 2000c UV/vis spectrophotometer (Peqlab Biotechnology) and RNA was stored at -80°C. To perform the cDNA synthesis, 100 ng of total RNA were used with Superscript III Reverse Transcriptase (Life Technologies) according to the protocol of Chen et al.[20] and primers as follows: miR-20a-3p stem-loop: 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTTAA-3', miR-20a-5p stem-loop: 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3', U6: 5'-GTCATCCTTGCGCAGG-3'.
Quantification of miR-20a-3p and miR-20a-5p by polymerase chain reaction (PCR)
Quantification of microRNAs was performed on a CFX96 Real-Time System (Bio-Rad, München, Germany) with SensiMix SYBR hi-ROX Kit (Bioline, Luckenwalde, Germany). Sequences of primers specific for human and murine sequences were as follows: miR-20a-3p forward: 5’- GCCCGCACTGCATTATGAGCACTTAAAG-3’; miR-20a-5p forward: 5’-GCCCGCTAAAGTGCTTATAGTGCAG-3’; universal reverse primer (used for miR-20a-3p and miR-20a-5p): 5'-GTGCAGGGTCCGAGGT-3'; U6 forward: 5'-CGCTTCGGCAGCACATATAC-3'; U6 reverse: 5'-AGGGGCCATGCTAATCTTCT-3'. After initial denaturation (95°C for 5 min) 45 amplification cycles were performed (95 ̊C for 10 s, 60 ̊C for 20 s, and 72 ̊C for 10 s), followed by a melting-curve analysis. microRNA signals were standardized to U6 RNA as reference.
Cloning of DNA plasmide pmiR-20a-3p and pmiR-20a-5p
cDNA sequences encoding for mature miR-20a-3p and miR-20a-5p mimicking small hairpin RNA (pmiR-20a-3p, pmiR-20a-5p) were cloned into the pSUPERIORpuro vector (OligoEngine, Seattle, WA, USA). The oligonucleotides miR-20a-3p oligonucleotide 1: 5’-gatccccACTGCATTATGAGCACTTAAAGttcaagagaCTTTAAGTGCTCATAATGCAGTttttta-3’ and miR-20a-3p oligonucleotide 2: 5’-tcgataaaaaACTGCATTATGAGCACTTAAAGtctcttgaaCTTTAAGTGCTCATAATGCAGTggg-3’ and the oligonucleotides miR-20a-5p oligonucleotide 1: 5’-gatccccTAAAGTGCTTATAGTGCAGGTAGttcaagagaCTACCTGCACTATAAGCACTTTAttttta-3’ and miR-20a-5p oligonucleotide 2: 5’-tcgataaaaaTAAAGTGCTTATAGTGCAGGTAGtctcttgaaCTACCTGCACTATAAGCACTTTAggg-3’, respectively, were hybridized by a temperature gradient (95°C to 4°C in 40 min). Due to the asymmetrical design of both complementary oligonucleotides, the hybridization products formed defined single stranded 5' overhangs for ligation into the BglII/XhoI (Thermo Scientific, Waltham, MA, USA) digested pSUPERIORpuro vector. After following ligation (T4 DNA Ligase; Thermo Scientific), positively selected clones were verified by restriction analysis and sequencing.
Transfection experiments
HaCat and B16 cells were transiently transfected with the microRNA mimicking vectors miR-20a-3p and miR-20a-35, respectively. Transfection experiments were performed using Lipofectamine2000 reagent (Life Technologies) according to the manufacturer's protocol.
Statistics
For data evaluation, the graphics and statistics software Graph Pad Prism V 5.01 (GraphPad Software, La Jolla, CA, USA) was used. Results of at least four experiments were statistically analyzed, using the unpaired Student‘s t-test, and expressed as the mean ±SD compared to control cells. Results of p≤0.05 (*), p≤0.01 (**), and p≤0.001 (***) were given as significance.
Suppressed expression of miR-20a-3p and miR-20a-5p correlates with with enhanced growth rate of skin tumor cells
Cellular proliferation analysis over a period of 144 h (Figure 1) showed reduced cell growth characteristics of non-malignant HaCat cells (24 h: 3.9x104±1.2x104; 48 h: 4.5x104±3.5x104; 72 h: 1.1x105±2.4x104; 96 h: 2.9x105±9.2x104; 120 h: 4.7x105±1.9x105; 144 h: 7.9x105±9.3x104) compared to malignant B16 cells (24 h: 2.4x104±4.3x103, p=0.0593; 48 h: 1.0x105±3.8x104, p=0.0815; 72 h: 4.0x105±9.0x104, p=0.0009; 96 h: 8.8x105±1.8x105, p=0.0012; 120 h: 1.6x106±2.3x105, p=0.0002; 144 h: 1.8x106±7.7x104, p<0.0001).
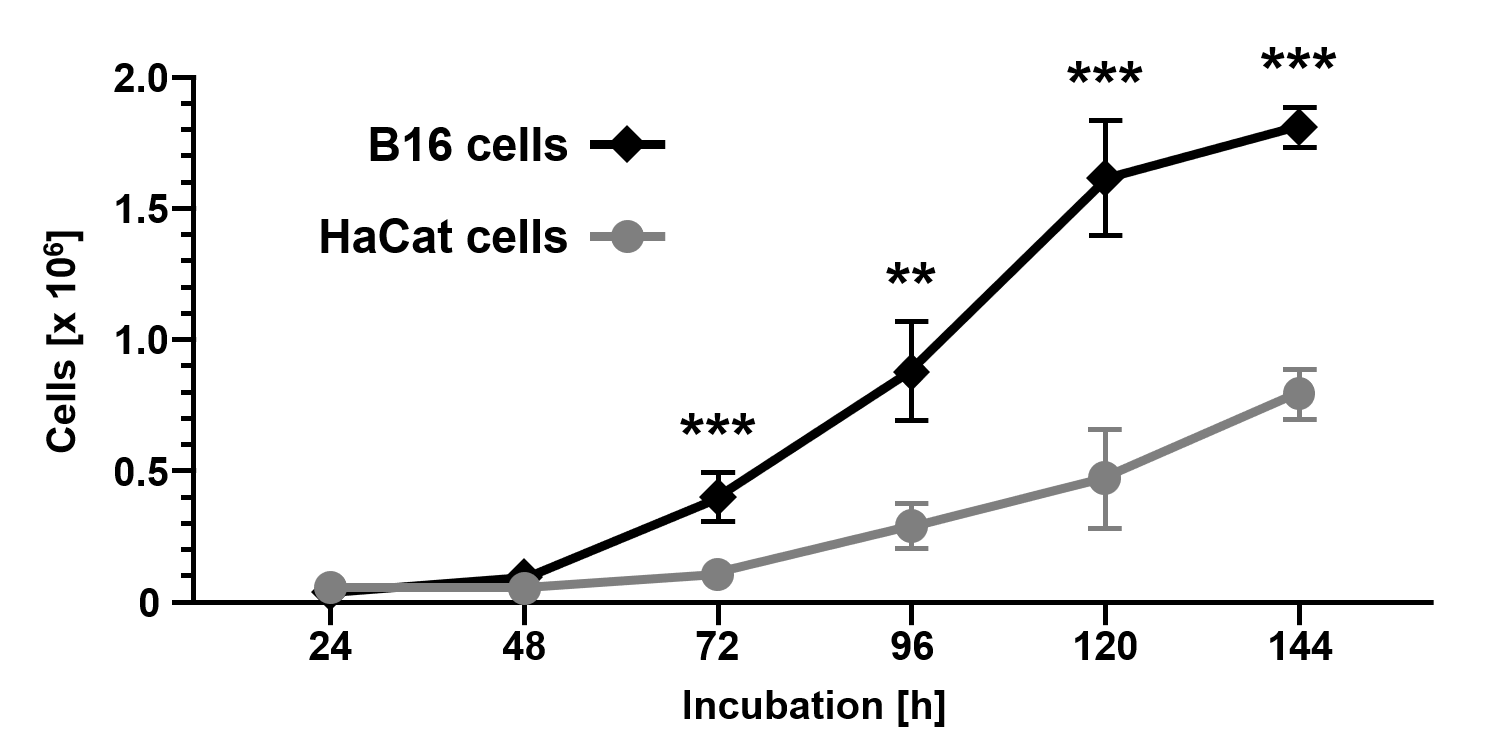
Figure 1. Cell growth of the melanoma cell line B16 compared to the non-malignant keratinocyte cell line HaCat. Cells were counted using a CASY Cell Counter and Analyzer Model TT (Roche Applied Science) at indicated time points. Data are given as the mean ± SD with p≤0.01 (**), and p≤0.001 (***) as determined by Student's t-test.
Subsequent detection and quantification of miR-20a-3p and miR-20a-5p demonstrated the reduced expression of both miRs in B16 cancer cells (miR-20a-3p: 1.7-fold±0.3-fold reduction, p=0.0381; miR-20a-5p: 1.7-fold±0.2-fold reduction, p=0.0015) compared to non-malignant HaCat cells (Figures. 2A and 2B). In solid tumors, microRNA's downregulation frequently point to tumor suppressive and anti-oncogenic properties of these miR species[21].
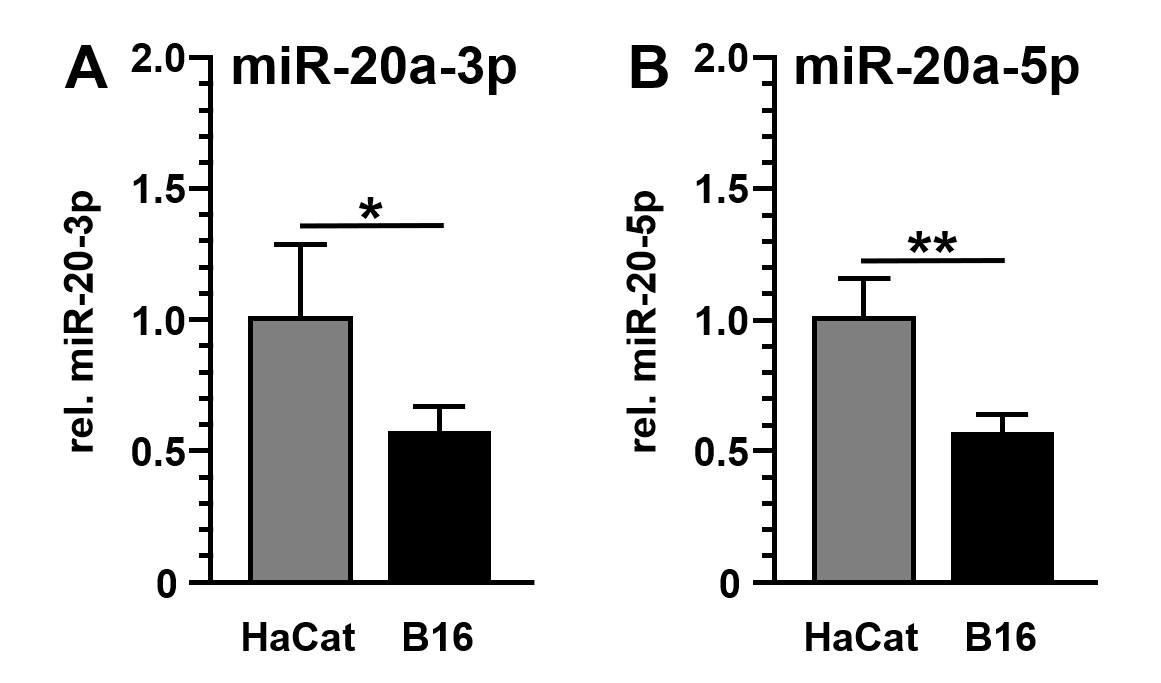
Figure 2. Expression of miR-20a-3p and miR-20a-5p in B16 and HaCat cells. MicroRNA levels of miR-20a-3p (A) and miR-20a-5p (B) were analyzed by quantitative reverse transcription and polymerase chain reaction (qRT-PCR) and standardized to U6 RNA expression levels. Data were given as the mean ± SD and expressed as fold change (HaCat = 1.0).
Overexpression of miR-20a-3p and miR-20a-5p inhibits cellular growth of skin tumor cells
miR-20a-3p and miR-20a-5p were cloned into the eukaryotic RNA expression vector pSUPERIORpuro (Figures 3A and 3B) and applied in transfection experiments. Transfection of the empty control vector led to slightly reduced cell growth of B16 cells (24 h: 8.4x103±7.9x103; 48 h: 1.6x104±8.2x103; 72 h: 6.6x104±3.5x104; 96 h: 3.1x105±8.2x104; 120 h: 6.0x105±2.5x105; 144 h: 9.0x105±1.8x105; Fig. 4) compared to untransfected cells (Figure 1). Transient overexpression of recombinant miR-20a-3p and miR-20a-5p revealed a minor but measurable attenuation of B16 cell growth in tendency (pmiR-20a-3p: 24 h: 7.1x103±6.8x103, p=0.7773; 48 h: 2.4x104±1.6x104, p=0.2554; 72 h: 6.8x104±5.4x104, p=0.9315; 96 h: 2.0x105±1.7x105, p=0.2942; 120 h: 3.5x105±3.1x105, p=0.2328; 144 h: 5.9x105±3.6x105, p=0.1529; pmiR-20a-5p: 24 h: 7.5x103±5.1x103, p=0.8260; 48 h: 1.7x104±9.2x103, p=0.8283; 72 h: 6.5x104±4.1x104, p=0.9648; 96 h: 2.4x105±1.7x105, p=0.4824; 120 h: 4.5x105±2.4x105, p=0.8239; 144 h: 6.7x105±1.4x105, p=0.9837; Figure 4).
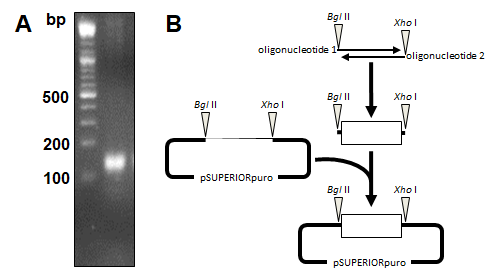
Figure 3. Cloning of miR20a-3p and miR-20a-5p encoding plasmid vector. (A) Oligonucleotides encoding for the miR20a-3p and miR-20a-5p small hairpin RNAs were hybridized forming single-stranded overhangs similar to BglII and XhoI endonuclease digestion. (B) The hybridization product of 160 base pairs (bp) was ligated into the BglII/XhoI digested vector pSUPERIORpuro (OligoEngine) and verified by sequencing.
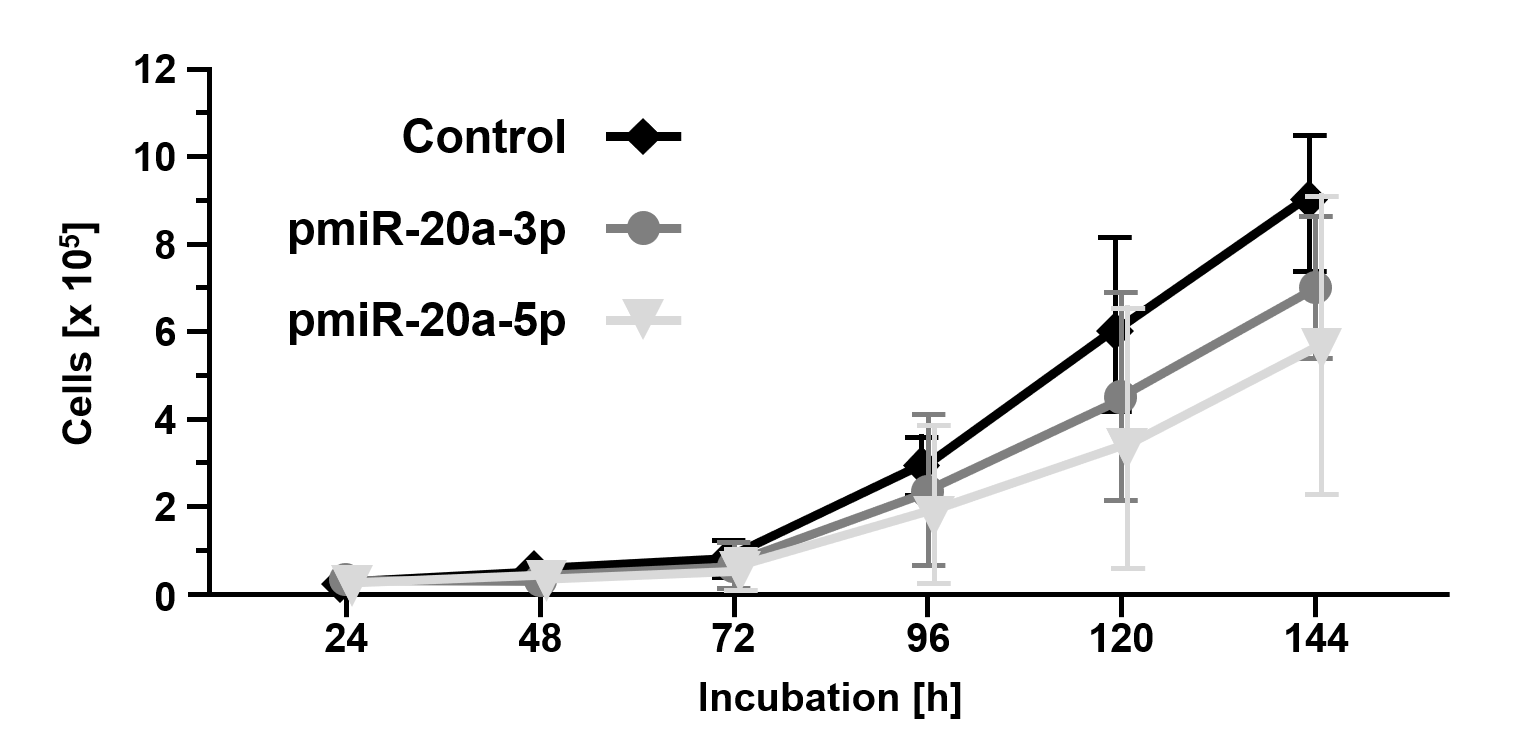
Figure 4. Overexpression of miR20a-3p and miR-20a-5p inhibits cellular growth of B16 tumor cells. Cell growth of B16 cells overexpressing miR20a-3p and miR-20a-5p compared to mock transfected B16 cells (control). Cells were counted using a CASY Cell Counter and Analyzer Model TT (Roche Applied Science) at indicated time points. Data are given as the mean ± SD.
Cells have to satisfy several requirements to become malignant, elegantly grouped by Hanahan and Weinberg as: enhanced cell motility, escalated angiogenesis, suppressed apoptosis, limitless replication, independence in growth signals, and resistance to anti-growth signals[22]. Dysregulation in the cellular safeguard machinery has a broad impact on cancer initiation and subsequent progression. In tumor biology, many miRs have been identified as crucial modulators of cancer malignancy usually classified in anti-oncogenic microRNAs (tumor suppressors) and pro-oncogenic microRNAs (oncomirs)[23]. Although miR-20a-3p's and miR-20a-5p's regulatory properties have been characterized in various tumor entities, surprisingly, nothing is known about their role in skin cancer development.
In the study presented here, expression and functionality of both the 3p and the 5p strand processed from the miR-20a precursor have been evaluated in an established skin cancer model for the first time. Both miR-20a forms were determined to fulfill a biological function. The expression of miR-20a-3p/miR-20a-5p was suppressed in melanoma cells B16 compared to the expression in non-malignant HaCat keratinocytes. Moreover, restoration of miR-20a-3p/miR-20a-5p levels in malignant B16 cells clearly demonstrated anti-proliferative properties of both and thus specifying miR-20a-3p/miR-20a-5p as tumor suppressors. Due to plasmid DNA transfer by electroporation is highly variable[24], however, transfection experiments overexpressing miR20a-3p/miR-20a-5p failed to become statistically significant.
Our findings are very similar to a study of Ottman et al., in which they showed downregulation and anti-proliferative properties of miR-20a in prostate cancer cells[25]. On the other hand, several studies demonstrated an upregulated expression of oncogenic miR-20a in other malignancies, highlighting miR-20a's heterogeneity in expression and functionality in a tissue and cell type-specific manner[26]. These conflicting data may be explained by the common mode of microRNA synthesis. Numerous microRNA genes are organized in gene clusters in which multiple microRNAs are expressed from a single promoter[27]. The resulting primary polycistronic transcript is subsequently processed into several individual microRNAs. The microRNA cluster miR-17 ~ 92 represents a microRNA cluster encoding for the microRNAs miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92a[28]. Abasi et al. examined the precursor and mature levels of miR-17, miR-20a, and miR-92a from the miR-17 ~ 92 cluster in various cancer cell lines and found cancer cell-specific variations of the three microRNAs compared to the level of unprocessed polycistronic precursor RNA[29].
In conclusion, our data suggest that melanoma cells may belong to the group of cancer cells in which miR-20a-3p/miR-20a-5p inhibit cell growth. Furthermore, during characterization of miR-20a-3p's/miR-20a-5p's functionality in cancer cells the role of other microRNAs encoded by the miR-17 ~ 92 microRNA cluster should not be ignored.
Author Contributions
HA, GD, EG, and MP conceived and designed the experiments. HA, EG and MP performed all experiments and GD, AM, MB, and MBS participated in analysis of the data. AM, and MBS wrote the paper; all authors read and approved the final manuscript.
Abbreviations
microRNA-20a (miR-20a), ethylenediaminetetraacetic acid (EDTA), polymerase chain reaction (PCR)
Latchana N, Ganju A, Howard JH, Carson and William E: MicroRNA dysregulation in melanoma. Surg Oncol 25: 184–9, 2016. PMid:27566021
View Article PubMed/NCBIBanerjee N, Bandyopadhyay AK, Dutta S, Das JK, Roy Chowdhury T, Bandyopadhyay A and Giri AK: Increased microRNA 21 expression contributes to arsenic induced skin lesions, skin cancers and respiratory distress in chronically exposed individuals. Toxicol 378: 10–6, 2017. PMid:28069514
View Article PubMed/NCBIYang CH, Yue J, Pfeffer SR, Handorf CR and Pfeffer LM: MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem 286: 39172–8, 2011. PMid:21940630 PMCid:PMC3234742
View Article PubMed/NCBIChen X, Li X and Qin Z: MicroRNA-21 promotes the proliferation and invasion of cholesteatoma keratinocytes. Acta Otolaryngol 136: 1261–6, 2016. PMid:27376830
View Article PubMed/NCBIWang J, Qiu Y, Shi N-W, Zhao J-N, Wang Y-C, Jiang H and Qian H-B: microRNA-21 mediates the TGF-beta1-induced migration of keratinocytes via targeting PTEN. Eur Rev Med Pharmacol Sci 20: 3748–59, 2016. PMid:27735045
PubMed/NCBIAgrawal R, Pandey P, Jha P, Dwivedi V, Sarkar C and Kulshreshtha R: Hypoxic signature of microRNAs in glioblastoma: Insights from small RNA deep sequencing. BMC Genomics 15: 686, 2014. PMid:25129238 PMCid:PMC4148931
View Article PubMed/NCBIZhi F, Shao N, Wang R, Deng D, Xue L, Wang Q, Zhang Y, Shi Y, Xia X, Wang S, Lan Q and Yang Y: Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro Oncol 17:383–91, 2015. PMid:25140035 PMCid:PMC4483096
View Article PubMed/NCBISi W, Shen J, Du C, Chen D, Gu X, Li C, Yao M, Pan J, Cheng J, Jiang D, Xu L, Bao C, Fu P and Fan W: A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ doi: 10.1038/cdd.2017.1762, 2017.
Calvano Filho CMC, Calvano-Mendes DC, Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR and Baracat EC: Triple-negative and luminal A breast tumors: Differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol 35: 7733–41, 2014. PMid:24810926
View Article PubMed/NCBIChen GS, Zhou N, Li J-Q, Li T, Zhang Z-Q and Si Z-Z: Restoration of miR-20a expression suppresses cell proliferation, migration, and invasion in HepG2 cells. Onco Targets Ther 9: 3067–76, 2016. PMid:27313460 PMCid:PMC4892835
PubMed/NCBIZhao S, Yao D, Chen J, Ding N and Ren F: MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS ONE 10: e0120905, 2015. PMid:25803820 PMCid:PMC4372287
View Article PubMed/NCBIPu Y, Yi Q, Zhao F, Wang H, Cai W and Cai S: MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int 16: 64, 2016. PMid:27499703 PMCid:PMC4974744
View Article PubMed/NCBIZhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W and Wang T: miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol Med Rep 14: 1742–50, 2016. PMid:27357419
View Article PubMed/NCBIWen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, Han J, Wei Q, Shen H, Sun B and Hu Z: Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer 137: 1679–90, 2015. PMid:25845839
View Article PubMed/NCBIYang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, Xu H, Zhong J and Zeng X: Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep 6: 429–34, 2017. PMid:28413641 PMCid:PMC5374949
View Article PubMed/NCBIBoelsma E, Verhoeven MC and Ponec M: Reconstruction of a human skin equivalent using a spontaneously transformed keratinocyte cell line (HaCaT). J Invest Dermatol 112: 489–98, 1999. PMid:10201534
View Article PubMed/NCBIStark H-J, Szabowski A, Fusenig NE and Maas-Szabowski N: Organotypic cocultures as skin equivalents: a complex and sophisticated in vitro system. Biol Proced Online 6: 55–60, 2004. PMid:15103399 PMCid:PMC389904
View Article PubMed/NCBIWang Q, Ilves H, Chu P, Contag CH, Leake D, Johnston BH and Kaspar RL: Delivery and inhibition of reporter genes by small interfering RNAs in a mouse skin model. J Invest Dermatol 127: 2577–84, 2007. PMid:17522708
View Article PubMed/NCBITurnbull DH, Ramsay JA, Shivji GS, Bloomfield TS, From L, Sauder DN and Foster FS: Ultrasound backscatter microscope analysis of mouse melanoma progression. Ultrasound Med Biol 22: 845–53, 1996. 00107-X
View ArticleChen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ and Guegler KJ: Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179, 2005. PMid:16314309 PMCid:PMC1292995
View Article PubMed/NCBIWeiss M, Brandenburg L-O, Burchardt M and Stope MB: MicroRNA-1 properties in cancer regulatory networks and tumor biology. Crit Rev Oncol Hematol 104: 71–7, 2016. PMid:27286699
View Article PubMed/NCBIHanahan D and Weinberg RA: Hallmarks of cancer: the next generation. Cell 144: 646–74, 2011. PMid:21376230
View Article PubMed/NCBIMarkopoulos GS, Roupakia E, Tokamani M, Chavdoula E, Hatziapostolou M, Polytarchou C, Marcu KB, Papavassiliou AG, Sandaltzopoulos R and Kolettas E: A step-by-step microRNA guide to cancer development and metastasis. Cell Oncology (Dordr) 40: 303–39, 2017. PMid:28748501
View Article PubMed/NCBIPotter H, Weir L and Leder P: Enhancer-dependent expression of human kappa immunoglobulin genes introduced into mouse pre-B lymphocytes by electroporation. Proc Natl Acad Sci U S A 81: 7161–5, 1984. PMid:6438633
View Article PubMed/NCBIOttman R, Levy J, Grizzle WE and Chakrabarti R: The other face of miR-17-92a cluster, exhibiting tumor suppressor effects in prostate cancer. Oncotarget 7: 73739–53, 2016. PMid:27650539 PMCid:PMC5340125
View Article PubMed/NCBIYang SS and Warner HR: The underlying, cellular and immunological factors in cancer and aging. Springer, New York, Heidelberg, ISBN 9781461362708, 1993.
View ArticleAltuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H: Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33: 2697–06, 2005. PMid:15891114 PMCid:PMC1110742
View Article PubMed/NCBIConcepcion CP, Bonetti C and Ventura A: The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J 18: 262–7, 2012. PMid:22647363 PMCid:PMC3592780
View Article PubMed/NCBIAbasi M, Kohram F, Fallah P, Arashkia A, Soleimani M, Zarghami N and Ghanbarian H: Differential Maturation of miR-17 ~ 92 Cluster Members in Human Cancer Cell Lines. Appl Biochem Biotechnol 182: 1540–7, 2017. PMid:28247308
View Article PubMed/NCBI