Abstract
Staphylococcus aureus is an important human pathogen and commensal, where the human nose is the predominant reservoir. To better understand its behavior in this environmental niche, RNA was extracted from the anterior nares of three documented S. aureus carriers and the metatranscriptome analyzed by RNAseq. In addition, the in vivo transcriptomes were compared to previously published transcriptomes of two in vitro grown S. aureus strains. None of the in vitro conditions, even growth in medium resembling the anterior nares environment, mimicked in vivo conditions. Survival in the nose was strongly controlled by the limitation of iron and evident by the expression of iron acquisition systems. S. aureus populations in different individuals clearly experience different environmental stresses, which they attempt to overcome by the expression of compatible solute biosynthetic pathways, changes in their cell wall composition and synthesis of general stress proteins. Moreover, the expression of adhesins was also important for colonization of the anterior nares. However, different S. aureus strains also showed different in vivo behavior. The assessment of general in vivo expression patterns and commonalities between different S. aureus strains will in the future result in new knowledge based strategies for controlling colonization.
Similar content being viewed by others
Introduction
Staphylococcus aureus is recognized as a major human pathogen, but also described as a human commensal. The human nose is its major reservoir and ecological niche where approximately 20–30% of humans are reported to be permanent carriers1. Even though carriage is usually asymptomatic, nasal carriage has a crucial function as a source of invasive infections in both community and hospital settings2. In recent decades, the prevalence of methicillin-resistant S. aureus (MRSA) has increased markedly3,4 and vancomycin resistant clones have also been reported5. The reasons for the rapid emergence of MRSA are still unclear and the spread cannot be explained solely be the antibiotic selection pressure6. In how far the natural ecological niche constitutes a selective advantage for MRSA remains to be elucidated. S. aureus carriage is influenced by host and environmental factors7 and also by interactions with other community members within the anterior nares8,9. Thus, it is assumed that a better understanding of the ecology of this niche may support strategies for limiting the carriage of S. aureus8,10,11.
S. aureus clearly belongs to the most studied bacterial species and its behavior under different in vitro conditions has been analyzed in great detail, where virulence factors12,13, global regulation14,15, stress response16, nutrient acquisition17, immune evasion18 and attachment mechanisms12, among others have been described. However, with the emergence of high throughput sequencing methods, the interest to understand the in vivo situation and the interactions between the host and the inhabiting bacterial communities has increased significantly19,20 and recent studies using deep sequencing transcriptomic analysis (RNAseq) indicate the behavior of S. aureus in vivo to be significantly different from that observed by the classical in vitro approach20,21.
Efforts to understand the survival and persistence of S. aureus in its ecological niche on the one hand aimed to understand the metabolic challenges. The analysis of nasal secretions resulted in the development of a synthetic nasal medium, which should mimic the nasal environment22. Thereby, iron limiting conditions as well as the absence of distinct amino acids were identified as shaping the transcriptional response. However, even though the use of such a medium may offer important insights into the global reaction of S. aureus to its natural conditions, it evidently cannot simulate the host environment and simulate e.g. adhesion processes19,23. Moreover the anterior nares are a complex ecosystem, which is characterized not only by the interaction of S. aureus with the host, but by the presence of a complex microbial community8,10 which may compete for nasal nutrients and interact with S. aureus. All these interactions cannot be easily mimicked in vitro.
In the current report, we analyzed the genome-wide transcriptional activity of S. aureus in vivo in the anterior nares of 3 volunteers at two different time points by RNAseq and compared the transcriptome profiles to those previously obtained by two distinct S. aureus strains under in vitro conditions24. The results of our study provide the first high-resolution analysis of the transcriptional response of S. aureus to its natural environment and will allow an understanding of how S. aureus persists in such a hostile environment niche.
Results and Discussion
The bacterial community of three S. aureus carriers
To gain insights into the behavior of S. aureus in its natural ecological niche, three healthy volunteers persistently colonized by S. aureus were selected. Volunteer 1 was colonized by a novel strain (novel spa type with repeat succession 15-21-12, S. aureus D1), volunteer 2 by a strain typed as spa type t0254 (S. aureus O1) and volunteer 3 by a strain typed as spa type t12 (S. aureus R1). Spa type t12 is reported to be one of the most abundant types colonizing healthy individuals25, specifically in young adults as is the case here. Multilocus sequence typing identified S. aureus D1 and R1 to belong to sequence type ST30 and S. aureus O1 to belong to sequence type ST15.
The bacterial communities of the anterior nares of these carriers were sampled during both winter (W) and summer (S) and the composition of the active bacterial community analyzed by deep sequencing of 16S rRNA amplicons following reverse transcription–polymerase chain reaction (RT–PCR) of total RNA extracts (Fig. 1, right). Analysis revealed a high abundance of S. aureus sequence reads in 5 of the 6 samples, and only in volunteer 1 it comprised <1% of sequence reads during summer (Volunteer 1S). A specifically high amount of sequence reads originating from S. aureus was observed in volunteer 3 (38–44%) indicating this organism to be specifically active in this person.
Description of the microbial community of three volunteers at two different time points during the year.
The graphic show the relative abunance per taxonomic group. The right side shows the abundance of species based on the sequencing of 16S rRNA amplificons (V1-V2), whereas the left side shows the abundance based on the amount of metatranscriptomic reads assigned to this species.
Overall, all three volunteers showed the presence of a bacterial community composed of core nasal colonizers such as Corynebacterium accolens/tuberculostearicum (19–42% relative abundance), Staphylococcus epidermidis/capitis/caprae (6–23%), Propionibacterium acnes (2–15%) or Peptoniphilus sp. (3–9%) among others (Fig. 1 right and Supplementary Dataset S1). While the composition of the bacterial communities of volunteer 2 and 3 were very similar during both sampling times (Fig. 1 right), evident differences were observed for volunteer 1, where the winter sampling showed a mild colonization by S. aureus (9%), whereas the summer community was dominated by Dolosigranulum pigrum and Corynebacterium propinquum/ pseudodiphtheriticum.
Metatranscriptomic analysis of nasal microbial communities
The same six in vivo samples described above were subject to a metatranscriptomic analysis by RNAseq. After quality filtering, 9–106 million sequence reads per sample were obtained (Table 1). Between 15 and 96% of those reads could be assigned to human RNA and were removed. After removal of 16S rRNA reads between 2 and 8 million of sequence reads remained as potential bacterial mRNA (Table 1).
A crucial step for evaluating the activity of S. aureus in its natural niche is the capability to separate sequence reads originating from S. aureus and S. epidermidis. Using a database of 25 S. aureus and 2 S. epidermidis genomes to assign reads to these species, between 12,000 and 120,000 reads showed a high similarity of (alignment length) * (% identity/ query length) ≥80% with S. aureus genomic sequences. However, of those reads between 35 and 95% could also be assigned to S. epidermidis, showing the similarity between the two species and the high level of possible mis-assignment. In contrast, using a similarity ratio ≥90%, the percentage of reads that could not be clearly assigned to one of those species decreased substantially to 9–34%. Only in case of sample 1S, 90% of the reads remained of unclear assignment (Table 1). This corresponds to the high relative abundance of S. epidermidis compared to that of S. aureus in this specific sample, as indicated by amplicon sequencing (Fig. 1 right). Thus, assignment of reads to S. aureus and S. epidermidis was performed using a similarity ratio ≥90% as cut-off, whereas assignment to bacterial genera was performed with a ≥80% similarity ratio as cut-off (Table 2).
In total, between 18,000 and 310,000 reads could be assigned to the different bacterial genera and species, which is only a subset of the metatranscriptomic reads remaining after depletion of human and ribsosomal reads (Table 2). The remaining unassigned mRNA reads illustrate the complexity of the anterior nare niche, and may represent transcripts pertaining to other community species such as eukaryotic species like fungi and protozoa26, although identifying these taxa was beyond the scope of this work. However, previous experiences also have shown an association between the sample quality and the number of RNAseq mapped reads obtained from it, i.e. samples with lower quality, and therefore more degraded RNA molecules, as present in in vivo samples yield both lower numbers of reads that can be assigned and reads mapped to genes27 and may explain why only a subset of metatranscriptomic reads could in fact be assigned here.
Comparison of the composition across the six nasal microbial communities under study as deduced by amplicon sequencing (Fig. 1 right) versus the composition as indicated by the relative amounts of transcripts assigned to the different species and genera (Fig. 1 left) indicated similar structures. This in turn indicates that representative amounts of metatranscriptomic reads were sampled in each case.
Potential S. aureus reads were then mapped against a database of orthologous groups of proteins previously built on the basis of 25 reference genomes24 and 73–80% of the reads could be successfully mapped to the S. aureus gene catalogue (Table 1). The remaining reads matched intergenic regions that mainly reflect the sites of transcription initiation and different RNA species. Analysis of the sequence reads that could be mapped to S. aureus versus those that could be mapped to both S. aureus or S. epidermidis considering a ≥90% similarity ratio showed, that for some genes all reads fall into the last category, indicating the presence of highly conserved gene regions, preventing a clear assignment (see Supplementary Dataset S2). As sample 1S mainly contains Staphylococcus sequence reads, which could not clearly be assigned, it was excluded from further detailed analysis keeping the samples in which S. aureus was shown to be active and abundant.
Functional categorization of transcripts expressed by S. aureus under in vivo conditions
Due to the difference in sequencing depth between the samples, random resampling was performed and the expression patterns analyzed on the basis of 5,000 sequence reads each (Supplementary Dataset S2 and Supplementary Fig. S1). These in vivo expression patterns were compared to those previously reported for S. aureus USA300 LAC and S. aureus IPL32 grown in vitro in rich medium (BHI) and a synthetic nasal medium (SNM)24 which mimics the nasal conditions22. Cluster analysis was used to determine the similarity between the global transcriptional profiles of S. aureus under the different conditions. The dendrogram (Fig. 2) shows that all 5 in vivo transcriptomes differed substantially from those of USA300 LAC and S. aureus IPL32 under in vitro growth conditions, that is, these two groups of samples were <50% similar, and those of both strains grown in synthetic nasal medium (SNM) were not more similar to the in vivo trancriptomes then those obtained in rich medium.
Cluster analysis within in vitro and in vivo conditions.
Dendrogram constructed by agglomerative hierachical clustering (group-average) based on a relative abundance matrix constructed from comparisons of metatransciptomic data between five in vivo samples analyzed here (depicted by filled squares) and in vitro transcriptomic data generated previously24 using S. aureus USA300 LAC and S. aureus IPL32 growing in BHI or SNM at different growth phases (exponential - Ex and stationary - St). The percentage similarity between conditions was calculated using the Bray-Curtis similarity algorithm.
It is important to note that the in vitro RNAseq libraries had been amplified using Ambion’s MessageAmp kit24, while the in vivo libraries in this work were generated using the Epicentre’s ScriptSeq Kit, but where isolation of RNA and sequencing were performed in the exact same manner. A comparison between these two methods on S. aureus cells grown to mid-log phase revealed only an additional 10% difference in the global expression profile compared to replicates (taking into account standardized read count across 2,582 S. aureus genes), where the rank-order of gene expression was mostly retained (Spearman rank correlation of 0.911) (data not shown).
Immense differences were obvious between the 5 in vivo transcriptomes sharing only >55% of similarity. Differences were observed not only between S. aureus transcriptomes from different individuals but also between transcriptomes obtained from the same volunteer at different timepoints, and thus from the same S. aureus strain. This reveals that differences in the transcriptomic profiles are not only due to the colonizing strain, but also to the environment provided by the host.
Gene expression analysis based on the relative abundances of gene transcripts assigned to their respective Clusters of Orthologous Groups (COG) showed no evident difference between in vivo and in vitro conditions (Supplementary Fig. S2), and mainly differences in the general distribution per categories between growth media were obvious, as previously reported24. However, differences were also visible in the relative transcript abundances of genes involved in inorganic ion transport and metabolism (category P) and secondary metabolites biosynthesis, transport and catabolism (category Q), where levels during growth in complex medium were lower compared to those during growth in either SNM or in vivo.
To obtain a better view on differences in transcriptomes, genes that were expressed to levels exceeding a normalized value of 1000 rpm after resampling were considered in more detail (Supplementary Dataset S2). Of the genes that were expressed in at least one in vivo sample at a level exceeding 1000 rpm, 240 genes were indicated to be expressed differently under in vivo versus in vitro conditions (see Materials and Methods and Supplementary Dataset S3).
Expression of genes encoding surface bound proteins
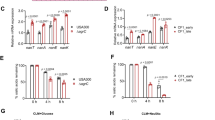
The adherence to components of the human extracellular matrix is a key step for the persistence of S. aureus in the nasal habitat and a total of 35 adhesins have been examined in detail28. Of these, the staphylococcal cell-wall protein clumping factor B (ClfB, USA300HOU_2630) which promotes adhesion to squamous epithelial cells29 was indicated as important for nasal colonization19. In fact, read counts indicating transcription of the encoding gene in vivo varied from 910–4660 reads per million of total sequence reads (rpm), whereas those from cells grown in SNM (in vitro) were only 330–550 rpm (Fig. 3A). Reads assigned to clfA (USA300HOU_0819) were similar between in vivo and in vitro growth in SNM. However, sdrC, D and E, all characterized as encoding crucial attachment factors13, but yet to be linked with nasal colonization were all expressed to a much higher extent in vivo compared to their previously reported in vitro expression (e.g. 340–1260 rpm in vivo versus ≤10 rpm in vitro for sdrC) (Fig. 3A). This indicated the importance of SdrCDE (USA300HOU_0555-0557) as attachment factors for nasal colonization. Furthermore, the sasF gene (USA300HOU_2646) was expressed to a much higher extend in vivo compared to in vitro (340–1330 rpm versus 14–120 rpm, Fig. 3A).
Comparison between the expression of various S. aureus survival factors.
Expression of genes encoding different adhesion factors (A), of genes involved in iron homeostasis (B), of genes involved in subversion of the host defense (C), of stress response genes (D), of genes encoding regulators (E) and methionine biosynthesis genes (F) of colonizing S. aureus strains in vivo (grey scaled) compared to their expression by S. aureus USA300 LAC or IPL32 during exponential growth in vitro on BHI or SNM as previously decribed24. Expression levels are given as rpm (reads per million) of total reads.
The Staphylococcus surface proteins can interact with different compounds in the extracellular matrix, and also the elastin-binding protein (ebpS, USA300HOU_1419) was expressed to a higher extent in vivo compared to its in vitro expression (350–1690 rpm in vivo versus 210–250 rpm after growth in SNM), whereas the cna gene (SAA6008_02751) encoding collagen adhesin was expressed only in two of the three volunteers (samples of volunteer 1 winter, 1W; and of volunteer 3 in summer and winter, 3S and 3W). Most intriguingly was the behavior of sasG (SACOL2505), which has been reported to promote adhesion to nasal epithelial cells13,30,31. This gene was expressed in vitro by the two strains previously analyzed only at levels <50 rpm of reads, but reached roughly 10,000 rpm in volunteer 2S (Fig. 3A). As mentioned above, the majority of transcripts obtained from sample 1S could not clearly be assigned to S. aureus or S. epidermidis, however, of the 583 reads that were mapped exclusively to S. aureus, >50% were due to transcription of sasG. Taken collectively, these levels of expression indicate that S. aureus uses a whole battery of adhesins to colonize the anterior nares, with the precise nature of the expression of adhesins being seemingly dependent on the colonizing strain and/or the human host.
Expression of genes involved in iron homeostasis
Much of the success of S. aureus as a major colonizer across the human body has been attributed to its capacity to retrieve iron from the host17,32 and iron limiting conditions as evidenced by the high expression of iron-regulated genes have recently been observed in culture media mimicking the conditions in the anterior nares22,24 or human plasma among others15. In fact, both the genes encoding proteins for the biosynthesis of staphyloferrrin B (sbnABCDEFGHI, USA300HOU_0127-0135) and staphyloferrin A (sfaCBAD, USA300HOU_2170-2173) as well as the respective transport systems (sirABC, USA300HOU_0126-0124; htsABC (USA300HOU_2169-2167). Similar to the previously described in vitro samples, expression of all four mentioned operons was high in all five in vivo samples indicating actual iron limitation governing the performance of S. aureus in vivo in the nares (see Fig. 3B and Supplementary Dataset S2).
High amounts of transcripts (up to 7060, 2740 and 5150 rpm, respectively in sample 3S) could be assigned to the sitADB (USA300HOU_0651-0653) iron transporter, however, similar high expression of this transporter has previously been observed during stationary phase of growth24. Transcription of various other transport systems was observed (fepABC, USA300HOU_0364-0366; fhuAB, USA300HOU_0668-0669; sstABCD, USA300HOU_0759-0762), however, only in the case of the fepABC iron transport system, an increased expression in vivo compared to in vitro exponential growth in complex medium of strains USA300 LAC and ILP32 could be observed (see Supplementary Dataset S2).
Genes of the isdBACDEF gene cluster (SACOL1138, USA300HOU_1064-1068) encoding iron regulated surface determinants were also previously reported as being upregulated by S. aureus in SNM22,24,32. Even higher expression of these genes was observed here in vivo and as an example, isdB transcripts accounted for 1200–4160 rpm in in vivo transcriptomes and only 60–90 rpm after growth of strains USA300 Lac or IPL32 in vitro in SMN (Fig. 3B).
Expression of genes required for subversion of the host defense
To survive within the host, S. aureus has developed various mechanisms to overcome the host immune defenses33. The staphylococcal complement inhibitor (SCIN), which interferes with all complement activation pathways is recognized as the most efficient complement inhibitor18 and the encoding scn gene has recently been shown to be highly expressed by S. aureus in an in vivo mouse model of infection20. During in vitro growth, its expression was typically relatively low and the level of transcripts never exceeded 1000 rpm24 (see also Fig. 3B) and low levels were also observed in a study comparing various in vitro conditions, where highest levels were observed during growth in human plasma15. Extreme differences were observed in the different volunteers and while in two volunteers expression levels were similar to those previously observed in vitro, in volunteer 3 transcript levels reached 22,000 rpm (Fig. 3B). Similarly the chp gene encoding chemotaxis inhibitory protein (USA300HOU_1947), where transcripts had been shown not to exceed 300 rpm in vitro24 was highly expressed in volunteer 3, reaching 13,600 rpm in sample 3W.
S. aureus can also secrete various exotoxins that damage the host cell plasma membrane, among them α-hemolysin, β-hemolysin, the bi-component leukocidins and γ-hemolysin and the phenol soluble modulins as well as δ-hemolysin34. While α- and β-hemolysin encoding genes (hly, USA300HOU_1099; hlb, SACOL2003) were expressed in vivo to a similar extent to previously described in vitro conditions, all of the in vivo samples (with the exception of 2S) showed a higher expression of γ-hemolysin encoding genes (hlgABC, USA300HOU_2402, 2405, 2404) and two of the volunteers (volunteer 2 and 3) showed expression of the δ-hemolysin encoding gene (hld, USA300HOU_2031), expression of which was not observed in vitro (Fig. 3C)24. δ-Hemolysin is encoded within RNAIII35 which is part of the agr regulon and belongs to a group of small peptide toxins known to activate, attract and lyse neutrophils, which comprise also the phenol soluble modulins (PSMs). Both volunteers 2 and 3 in addition to hld also expressed the gene encoding cytosolic toxin PSMα1 (see Fig. 3C). However, in contrast to the expression of PSMβ1 and PSMβ2 under in vitro conditions in nasal medium22 with amounts of 1550–1610 rpm and 1100–1270 rpm, respectively by S. aureus IPL32 and S. aureus USA300 LAC24, their expression in vivo was only observed in one volunteer at levels not exceeding 370 and 120 rpm, respectively (Fig. 3C). This observation is in accordance with previous analysis, where expression of PSMβ was observed only in vitro20. Among the bi-component toxins not only were hlgABC shown to be expressed in vivo, but high expression of a gene encoding a putative leucocydin (lukFS, USA300HOU_2011/USA300HOU_2013) specifically in volunteer 3 was observed, the importance of which remains to be elucidated.
Superantigens are virulent polypeptides that are capable of causing nonspecific T cell activation by circumventing normal antigen processing in the human host. The tst gene encoding toxic shock syndrome toxin-1 (TSST-1) has recently been observed in 15% of nasal S. aureus isolates from medical students in Poland36. Interestingly, the tst gene (SAV2011) was not only present in at least one volunteer, it was also expressed in this volunteer (volunteer 3) at levels reaching 1230 rpm (Fig. 3C).
Expression of genes involved in stress response
Reactive oxygen species are produced by bacteria as a by-product of aerobic growth but also by the host, specifically as part of an oxidative killing mechanism37. Protection of S. aureus from such oxidative stress is mediated by a battery of enzymes among them KatA catalase and AhpC alkylhydroperoxide reductase, which both have been described as essential for nasal colonization by scavenge exogenously or endogenously produced hydrogen peroxide38. Both katA (USA300HOU_1277) and ahpC (USA300HOU_0404) were expressed at high levels in vivo (>930 and >1320 rpm, see Fig. 3D) of one order of magnitude higher compared to their levels after growth of USA300 LAC or IPL32 in SNM (katA<370 rpm, ahpC <540 rpm), indicating S. aureus to experience oxidative stress in the anterior nares. Another gene highly expressed in vivo was asp23 (USA300HOU_2175, Fig. 3C). Asp 23 was initially described as a protein that accumulates after alkaline shock39 and has recently been identified as a membrane associated protein16 which may be important for cell envelope homoeostasis, specifically under adverse environmental conditions. In accordance with the previously observed high abundance of this protein in the complete proteome of S. aureus16, a high expression (480–1800 rpm), had been observed in vitro24, however, even when taking into account that 30–49% of reads could also be mapped to S. epidermidis (see Supplementary Dataset S2), higher expression levels were observed in vivo indicating Asp23 to be of major importance for survival of S. aureus in the host.
S. aureus has been shown to accumulate the compatible solute glycine betaine in response to osmotic stress40. A common bioysynthetic pathway for glycine betaine is its formation from choline, which in S. aureus is catalyzed by a choline dehydrogenase BetA forming glycine betaine aldehyde and a glycine betaine aldehyde dehydrogenase BetB forming glycine betaine41. Both betA and betB genes encoding the biosynthetic enzymes (USA300HOU_2605-2606) as well as betT encoding a choline transporter (USA300HOU_2610) were highly expressed in vivo, indicating S. aureus to experience osmotic stress in the nasal cavity (Fig. 3D).
CsbD is a bacterial general stress response protein, however, its role in stress response is unclear42. Typically, two csbD-like proteins are encoded in the genome of S. aureus strains, and both genes (USA300HOU_0868 and 1625) were expressed at levels under in vivo conditions two orders of magnitude higher (up to 17,640 and 25,860 rpm, respectively) compared to levels observed in vitro (<90 rpm) constituting two of the most abundant transcripts, indicating their importance for nasal colonization (Fig. 3D). Only 3–12% of the sequence reads attributed to csbD2 expression in S. aureus could not clearly be assigned and may also originate from S. epidermidis. Similarly a dps family protein (USA300HOU_2128), a family comprising proteins protecting DNA under starved conditions, was expressed to a much higher extent in vivo compared to in vivo (up to 11,970 rpm).
Evidently, also genes encoding various membrane proteins (SAV0374, SAV0574, SAV1030 and SAV1359) were extremely differently expressed under in vivo versus in vitro24 conditions (Supplementary Dataset S2 and Fig. 3D). A further protein where the encoding gene showed a 2 order of magnitude difference in expression was VraX (Fig. 3D). vraX expression was also previously shown to be upregulated by multiple cell wall and/or membrane active compounds43,44,45 indicating that vraX up-regulation follows all forms of cell membrane and/or cell wall metabolism insult and that the anterior nares are a harsh environment for bacteria to survive.
Expression of genes encoding regulatory systems
Regulation in S. aureus has been widely described in vitro14 and is performed by two-component regulatory systems (TCRS) and the SarA homologs, that commonly control many virulence factors46. Of the regulatory systems, substantial differences between in vivo and in vitro conditions were observed for the agrAC genes (USA300HOU_2035-2034) encoding the two-component AgrAC regulatory system as well as the sarA gene. While the agrAC genes represented 1070–4500 and 1170–5260 rpm, respectively, during growth of USA300 LAC and IPL32 strains in vitro24, they summed up to 510 and 860 rpm, respectively in vivo (Fig. 3E). Low expression of agr genes concomitantly with a high expression of virulence factors in vivo has been reported recently20 and some of the virulence factors assumed to be upregulated by the Agr or SarA system47, such as γ-hemolysin were in fact upregulated in vivo in the nose. Thus, these staphylococcal virulence factors may be under the control of additional regulatory mechanisms in vivo.
Expression of genes involved in metabolic processes
The regulation of a wide variety of virulence factors its dependent on the nutrient availability48 and nasal secretions were analyzed previously with the objective to determine the abundance and limitations of potential nutrients that could be consumed by nasal communities, specifically S. aureus22. Methionine biosynthesis had been reported to be crucial for S. aureus to inhabit the nasal habitat, and genes encoding methionine biosynthetic enzymes such as cystathionine-γ-synthase (metI) and cystathionine-β-lyase (metC), as well as two L-methionine ABC-transport systems were reported to be up-regulated during growth in synthetic nasal medium22,24 and MetI was even reported to be indispensable for growth in SNM. However, while the USA300HOU_0376-USA300HOU_0380 metEHCI methionine biosynthesis operon was highly expressed in SNM, it was only poorly expressed during growth in rich medium as well as in vivo (Fig. 3F). In addition the strong upregulation of two L-methionine ABC-transport systems (metN1P1Q1, SACOL0504-0506 and metN2P2Q2 USA300HOU_0847 – 0849) previously observed in SNM22,24 did not occur in vivo (Fig. 3F), indicating the strong upregulation of methionine biosynthetic genes and the respective transport systems to be a medium artifact.
Conclusions
Even though the epidemiology and characterization of strains inhabiting the anterior nares as well as the characterization of gene expression profiles under in vitro conditions mimicking those in vivo have established a sound knowledge base of S. aureus colonization strategies, the mechanisms used by S. aureus in vivo to survive in the nasal environment are still poorly understood19,49. The comparison between in vivo gene expression profiles across different human hosts and then comparing those with published in vitro expression profiles has shown that the host environment has a stronger impact on the expression profile than growth phase or medium composition, supporting the notion that understanding colonization by pathogenic or commensal microorganisms as well as of infection processes necessitates in vivo studies. The differences in gene expression profiles between samples has revealed diverse mechanisms used by S. aureus to persist in the human nose. As previously described19,22,32, survival in the nose is strongly controlled by the limitation of iron and evident by the expression of iron acquisition systems (fepABC, sirABC, htsABC). Besides the need to acquire iron, S. aureus cells clearly experience different environmental stresses, which they commonly attempt to overcome by the expression of compatible solute biosynthetic pathways, changes in the cell wall composition and the synthesis of general stress proteins. Moreover, the expression of adhesins is important for colonization of the nares as evident by the consistent expression of sdrCDE, clfB and ebpS in vivo. However, different S. aureus strains also showed different in vivo behavior, maybe also due to differences in the environment set by the host. This is most evident for adhesins, where sasG expression was dominant in only one volunteer. Overall the analysis of general expression patterns and commonalities between different S. aureus strains in distinct host environments can provide new knowledge to base strategies to combat colonization by S. aureus or other opportunistic pathogens.
Materials and Methods
Confirmation and characterization of persistent S. aureus carriers
Persistent colonization of putative S. aureus carriers was analyzed by inoculation of cotton swabs of the anterior nares of volunteers on selective chromogenic media (CHROMagarTM Staph aureus) and incubation for 48 h at 37 °C in ambient air. Suspicious colonies were confirmed as S. aureus by amplification of the nuc gene as previously described50. Isolated colonies from two nasal swabs from persistent carriers (those where S. aureus was observed at least four times in a one year period) were characterized by sequencing of the repeat region of the protein A gene (spa)51. The spa types were assigned through the Ridom web server (http://www.spaserver.ridom.de/). As from the different carriers always the same spa type was observed, further identification of a single isolate was performed by Multilocus Sequence Typing (MLST) as previously described52 and the sequence type assigned using the MLST database (http://saureus.mlst.net). Three male volunteers aged 30–50 years colonized in their anterior nares by S. aureus were thereby selected for the present study. Informed consent was obtained from all three volunteers included in this study for the analyses described. The study was approved by the Ethical Committee of the Medical Faculty of the University of Münster and of the Ärztekammer Westfalen-Lippe (file number 2010-468-f-S) and was carried out in accordance with the relevant guidelines.
Total RNA extraction, purification and mRNA enrichment
Samples for metatranscriptomic analysis were taken at two time points (February and August 2013) using dry sterile cotton swabs. As only small amounts of RNA could be obtained from a single nasal swab, RNA from consecutive swabs was pooled. To verify the stability of the microbial community over short period of times, single swabs were taken daily from both anterior nares of a volunteer and the microbial community structure profiled by T-RFLP (terminal restriction fragment length polymorphism) as previously described53. After confirming the stability of the community over short time frames, swabs were taken from both anterior nares of each volunteers in the morning and evening of five consecutive days. Samples were stored at −80 °C after addition of 100μl of RNA-later®.
Single samples (swabs) were resuspended in 1 ml of cold buffer RLT (Qiagen) supplemented with 1% β-mercaptoethanol (Sigma) and transferred to lysing matrix B tubes (MP Biomedicals) on ice. Samples were disrupted using the FastPrep-24® instrument (MP Biomedicals) at an intensity of 5.5 for 40 s. Samples were returned to ice for 4 min, disrupted further using the same settings and then centrifuged at 13 500 × g for 10 min at 4 °C to remove the cell debris. The supernatant was subsequently transferred to 1.5 ml RNase free Biopur™ centrifuge tubes (Eppendorf) and the RNA extracted using the RNeasy Mini Kit according to the manufacturer’s instructions, including optional DNase treatment on the column (Qiagen). RNA was eluted with nuclease-free water (Ambion), pooled and concentrated by ethanol precipitation using standard procedures.
Amplicon library preparation and analyses
An aliquot of each pooled RNA sample was transformed into cDNA using the QuantiTect® reverse Transcription kit (Qiagen) following the manufacturer instructions and 16S rRNA amplicon libraries comprising the V1-V2 variable regions were prepared as previously described54. These 6 samples were sequenced on the MiSeq instrument (Illumina) with processing of sequence reads performed as previously described54. In brief, quality filtered reads were trimmed conservatively to 80 nt and the paired ends matched to give 160 nt for downstream analysis. Reads were clustered allowing for two mismatches55 and the dataset then filtered to consider only those phylotypes that were present in at least one sample at a relative abundance >0.05% of the total sequences of that sample. The mean number of sequences per sample was 11345 +/− 2482 totaling 68072 sequence reads and the taxonomic affiliation of the 142 phylotypes was assigned as previously described54.
Transcript library construction and sequencing
mRNA was enriched from each of the 6 samples using Terminator™ 5′-Phosphate-Dependent Exonuclease (Epicentre) according to the manufacturer’s instructions. Samples were further concentrated by ethanol precipitation and re-eluted in nuclease-free water (Ambion). RNA integrity and quantity was measured using the Agilent 2100 Bioanalyzer (Agilent Technologies) and NanoDrop 1000 spectrophotometer (Thermo Scientific). Libraries were generated using the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre). For each library, 50 ng of enriched mRNA was used in each reaction according to the manufacturer’s instructions and the libraries purified using the Minelute PCR Purification Kit (Qiagen). The libraries were further purified for removal of potentially contaminating primer dimers by agarose gel electrophoresis and excision and purification of the 250–650bp fragments by the QIAquick Gel extraction Kit (Qiagen). Libraries were assessed for quality using the Agilent 2100 Bioanalyzer (Agilent Technologies) and were sequenced on the Illumina HiSeq 2500 platform using the TruSeq SR Cluster Kit v3-cBot-HS (Illumina). Four libraries were multiplexed per lane (12 pM/library) and sequenced to 200 cycles in both directions.
RNA-Seq data processing
Each library produced between 50 and 105 million reads, which were pre-processed for quality and trimmed using a combination of in-house Ruby scripts and open source tools (http://bioinformatics.ucdavis.edu/index.php/Trim.pl and http://www.mothur.org/)56. Reads were collapsed into representative reads using FASTX toolkit V. 0.0.13.2 collapser (http://hannonlab.cshl.edu/fastx_toolkit/).
Human associated RNAs were identified and removed from the in vivo sample using BLAT searches57 of an in-house database of the human genome repository (human RefSeq, chromosome records with gap adjusted concatenated NT_contigs) downloaded from the NCBI ftp site, blastdb (March 2013). Reads with alignment coverage (on the query) ≥30% were removed from the dataset. Ribosomal sequences contained within the datasets were detected with HMMER (version 3.058) using models based on multiple sequence alignments of the 5S, 16S and 23S rRNAs59 and by the use of riboPicker (version 0.4.3). Results of the two methods were compiled and used to depurate datasets.
Seven nucleotide sequence databases were constructed and used to assign metatranscriptomic reads. These databases comprised the complete genomes from 1) the 25 S. aureus strains24, 2) S. epidermidis strains ATCC12228 and RP62A, 3) F. magna (ATCC29328), 4) M. catarrhalis RH4, 5) P. acnes SK137, 6) D. pigrum ATCC51524, 7) Corynebacterium accolens ATCC49725 and Corynebacterium variabile DSM44702 and 8) Peptoniphilus duerdenii (ATCCBAA164). The metatranscriptomic datasets devoid of human and ribsosomal reads were individually analyzed using this set of blast databases. Reads were assigned to a genus, if the ratio (alignment length * % identity)/ query length) exceeds 80%. As a reasonable number of reads could not clearly be assigned to either S. aureus or S. epidermidis, a ratio (alignment length * % identity)/ query length) >90% was used to assign reads to these species (see Table 1).
Presumptive S. aureus reads were mapped against a previously described S. aureus OG/Position Specific Scoring Matrix (PSSM) database24 using rpstblastn. An alignment ratio of ≥60% (alignment length * % identity/ query length) was used for the positive assignment of sequences to the orthologous groups. Reads that mapped to S. aureus were further blasted (blastn) against two genomes of S. epidermidis (S. epidermidis strains ATCC12228 and RP62A). Reads that showed an alignment ratio ≥90% were aggregated and assigned as reads that could originate from either S. aureus or S. epidermidis (Supplementary Dataset S2).
As the number of S. aureus sequence reads varied between libraries (see Table 2) and then to compare with recently published in vitro data that comprised orders of magnitude more sequence reads24, mapped read counts were re-randomized to the smallest library size of 5000 reads using an in-house Perl script20. Finally all read count data were normalized to reads per million (rpm).
Clusters of Orthologous Groups (COGs) were assigned to the probable S. aureus reads by querying the COG profiles deposited in the Conserve Domain Database (CDD) database60 (downloaded from the NCBI ftp site/ little endian) using rpsblast (version 2.2.25). The results were parsed using an in-house Perl script.
Data analysis and interpretation
In order to compare the different in vivo S. aureus transcriptomic libraries with each other and then also with other published datasets (i.e. the in vitro libraries from Chaves-Moreno et al.24), a sample-similarity matrix was generated using the Bray–Curtis coefficient61 and gene-expression profiles compared using group-average agglomerative hierarchical clustering using PRIMER (v.6.1.6, PRIMER-E, Plymouth Marine Laboratory, Plymouth, UK)62.
Previous analysis of S. aureus 685020 and S. aureus SH1000 global transcriptomic profiles using the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (as performed here), showed similarities of 80–85% between replicates. Then, a direct comparison between the global transcriptomic profiles obtained from S. aureus SH1000 cells grown to mid-log phase in BHI using the MessageAmp™ II-Bacteria RNA amplification kit (Ambion) with a T7 primer modified to include a BpmI restriction site for removal of poly-A tails prior to sequencing as described by Chaves-Moreno et al.24 or the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre) (like the in vivo samples of this work) revealed global expression profiles of 75% similarity (using the Bray-Curtis algorithm) with a Spearman rank correlation of 0.911 (based on the read counts of the 2,582 S. aureus genes). So we can deduce that the choice of RNAseq library preparation method may contribute to an additional 10% difference in the global expression profile while retaining the rank-order of gene expression.
As the in vivo transcriptomes were very different and cannot be regarded as replicates, a detailed comparison with previous in vitro data24 was based on those genes that were expressed in at least one in vivo sample at a level exceeding 1000 rpm (Supplementary Fig. S1). Only genes where the expression level under at least one in vitro condition differed to that of the mean in vitro expression level of S. aureus USA300 strain LAC and S. aureus IPL32 growing in SNM or BHI, respectively, by at least one order of magnitude were considered for discussion in this paper (see Supplementary Dataset S3). Genes where the % of reads that also map to S. epidermidis exceeded 50% under any condition were not considered. Overall 240 genes were further compared.
Data accessibility
Sequences generated from these 6 in vivo samples were deposited in the NCBI Gene Expression Omnibus (GEO) repository under the accession number GSE73485.
Nucleotide sequences of all 142 phylotypes determined using Illumina-based amplicon deep-sequencing, their relative abundance (in%) and their phylogenetic assignment, an overview of the expression of S. aureus genes under in vivo conditions analyzed here compared to previously described in vitro conditions and an overview of genes where the expression level under at least one in vitro condition differed to that previously described in vitro are provided as Supplementary Dataset S1,S2.
Additional Information
How to cite this article: Chaves-Moreno, D. et al. Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci. Rep. 6, 33174; doi: 10.1038/srep33174 (2016).
References
van Belkum, A. et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol 9, 32–47 (2009).
von Eiff, C., Becker, K., Machka, K., Stammer, H. & Peters, G. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344, 11–16 (2001).
Rasigade, J.-P. et al. Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981–2007. J Infect Dis 201, 1589–1597 (2010).
Pfingsten-Würzburg, S., Pieper, D. H., Bautsch, W. & Probst-Kepper, M. Prevalence and molecular epidemiology of meticillin-resistant Staphylococcus aureus in nursing home residents in northern Germany. J Hosp Infect 78, 108–112 (2011).
Melo-Cristino, J., Resina, C., Manuel, V., Lito, L. & Ramirez, M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet 382, 205 (2013).
Rasigade, J.-P. & Vandenesch, F. Staphylococcus aureus: A pathogen with still unresolved issues. Infect Genet Evol 21, 510–514 (2014).
Kluytmans, J., van Belkum, A. & Verbrugh, H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10, 505–520, (1997).
Yan, M. et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14, 631–640 (2013).
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
Wos-Oxley, M. L. et al. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4, 839–851 (2010).
Faust, K. et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8, e1002606 (2012).
Corrigan, R. M., Miajlovic, H. & Foster, T. J. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9, 22 (2009).
Foster, T. J., Geoghegan, J. A., Ganesh, V. K. & Höök, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nature Rev Microbiol 12, 49–62, (2014).
Ibarra, J. A., Pérez-Rueda, E., Carroll, R. K. & Shaw, L. N. Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14, 126 (2013).
Mader, U. et al. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet 12, e1005962 (2016).
Müller, M. et al. Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol Microbiol 93, 1259–1268, (2014).
Hammer, N. D. & Skaar, E. P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol 65 (2011).
Rooijakkers, S. H. M. et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nature Immunol 6, 920–927 (2005).
Burian, M., Wolz, C. & Goerke, C. Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5, e10040 (2010).
Szafranska, A. K. et al. High-resolution transcriptomic analysis of the adaptive response of Staphylococcus aureus during acute and chronic phases of osteomyelitis. mBio 5, e01775–01714 (2014).
Cheung, A. L., Yang, S.-J., Bayer, A. S. & Xiong, Y. Q. Disparity in the in vitro versus in vivo regulation of fibronectin-binding proteins by 2 global egulators, saeRS and sigB, in Staphylococcus aureus. J Infect Dis 200, 1371 (2009).
Krismer, B. et al. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10, e1003862 (2014).
Burian, M. et al. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J Infect Dis 201, 1414–1421 (2010).
Chaves-Moreno, D. et al. Application of a novel “pan-genome”-based strategy for assigning RNAseq transcript reads to Staphylococcus aureus strains. PLoS ONE 10, e0145861 (2015).
Sangvik, M. et al. Age- and gender-associated Staphylococcus aureus spa types found among nasal carriers in a general population: the Tromsø Staph and skin study. J Clin Microbiol 49, 4213–4218 (2011).
Rivera, F. et al. Pathogenic and free-living protozoa cultured from the nasopharyngeal and oral regions of dental patients. Environ Res 33, 428–440 (1984).
Gallego Romero, I., Pai, A. A., Tung, J. & Gilad, Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC biology 12, 42 (2014).
Clarke, S. R. & Foster, S. J. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol 51, 187–224 (2006).
O’Brien, L. M., Walsh, E. J., Massey, R. C., Peacock, S. J. & Foster, T. J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4, 759–770 (2002).
Roche, F. M., Meehan, M. & Foster, T. J. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149, 2759–2767 (2003).
Corrigan, R. M., Rigby, D., Handley, P. & Foster, T. J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446 (2007).
Friedman, D. B. et al. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog 2, e87 (2006).
Foster, T. J. Immune evasion by staphylococci. Nature Rev Microbiol 3, 948–958 (2005).
Vandenesch, F., Lina, G. & Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol 2, 12 (2012).
Benito, Y. et al. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6, 668–679 (2000).
Piechowicz, L., Garbacz, K., Wiśniewska, K. & Dąbrowska-Szponar, M. Screening of Staphylococcus aureus nasal strains isolated from medical students for toxin genes. Folia Microbiol 56, 225–229 (2011).
Hampton, M. B., Kettle, A. J. & Winterbourn, C. C. Involvement of superoxide and myeloperoxidase in oxygen-dependent killing of Staphylococcus aureus by neutrophils. Infect Immun 64, 3512–3517 (1996).
Cosgrove, K. et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189, 1025–1035 (2007).
Kuroda, M., Ohta, T. & Hayashi, H. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem Biophys Res Commun 207, 978–984 (1995).
Graham, J. E. & Wilkinson, B. J. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol 174, 2711–2716 (1992).
Chen, C. et al. Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus. Appl Environ Microbiol 80, 3992–4002 (2014).
Prágai, Z. & Harwood, C. R. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148, 1593–1602 (2002).
Utaida, S. et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149, 2719–2732 (2003).
Pietiäinen, M. et al. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10, 429 (2009).
Dengler, V., Meier, P. S., Heusser, R., Berger-Bächi, B. & McCallum, N. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol 11, 16 (2011).
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H. & Xiong, Y.-Q. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Mic 40, 1–9 (2004).
Novick, R. P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48, 1429–1449 (2003).
Somerville, G. A. & Proctor, R. A. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev 73, 233–248 (2009).
Muthukrishnan, G. et al. Exoproteome of Staphylococcus aureus reveals putative determinants of nasal carriage. J Proteome Res 10, 2064–2078 (2011).
Brakstad, O. G., Aasbakk, K. & Maeland, J. A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol 30, 1654–1660 (1992).
Shopsin, B. et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 37, 3556–3563 (1999).
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J. & Spratt, B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38, 1008–1015 (2000).
Camarinha-Silva, A., Wos-Oxley, M. L., Jauregui, R., Becker, K. & Pieper, D. H. Validating T-RFLP as a sensitive and high-throughput approach to assess bacterial diversity patterns in human anterior nares. FEMS microbiology ecology 79, 98–108 (2012).
Camarinha-Silva, A. et al. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol 16, 2939–2952 (2014).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009).
Kent, W. J. BLAT–the BLAST-like alignment tool. Genome Res 12, 656–664 (2002).
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41, e121–e121 (2013).
Huang, Y., Gilna, P. & Li, W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25, 1338–1340 (2009).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5, R7 (2004).
Bray, J. R. & Curtis, J. T. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27, 325–349 (1957).
Clarke, K. R., Warwick, R. M. R. M. & Laboratory, P. M. Change in marine communities : an approach to statistical analysis and interpretation. 2nd ed edn, (Plymouth, U.K.: PRIMER-E Ltd, 2001).
Acknowledgements
The authors would like to thank Karsten Becker from the University Hospital Münster for collaboration in sampling, Robert Geffers and Michael Jarek from the Helmholtz Centre for Infection Research (HZI, Braunschweig) for sequencing support and Iris Plumeier (HZI, Braunschweig) for technical assistance. Financial support was provided by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung-BMBF) “Medizinische Infektionsgenomik” (grant 0315832B.).
Author information
Authors and Affiliations
Contributions
D.C.-M., A.P.A.O., E.M. and D.H.P. conceived and designed the experiments, D.C.-M. and A.P.A.O. performed the experiments, D.C.-M., M.L.W.-O., R.J. and D.H.P. performed the bioinformatic and statistical analyses, D.C.-M., M.L.W.-O. and D.H.P. wrote the manuscript. All the authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chaves-Moreno, D., Wos-Oxley, M., Jáuregui, R. et al. Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci Rep 6, 33174 (2016). https://doi.org/10.1038/srep33174
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep33174
This article is cited by
-
Exploring differentially expressed genes of Staphylococcus aureus exposed to human tonsillar cells using RNA sequencing
BMC Microbiology (2023)
-
Unravelling the physiological roles of mazEF toxin–antitoxin system on clinical MRSA strain by CRISPR RNA-guided cytidine deaminase
Journal of Biomedical Science (2022)
-
Iron-withdrawing anti-infectives for new host-directed therapies based on iron dependence, the Achilles’ heel of antibiotic-resistant microbes
Environmental Chemistry Letters (2021)
-
Antibiofilm potential of Psidium guajava and Passiflora edulis pulp extracts against Staphylococcus aureus, cytotoxicity, and interference on the activity of antimicrobial drugs
Future Journal of Pharmaceutical Sciences (2020)
-
Using Bacterial Transcriptomics to Investigate Targets of Host-Bacterial Interactions in Caenorhabditis elegans
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.