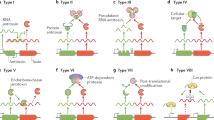
Abstract
Bacterial persister cells constitute a subpopulation of genetically identical, metabolically slow-growing cells that are highly tolerant of antibiotics and other environmental stresses. Recent studies have demonstrated that gene loci known as toxin-antitoxin (TA) modules play a central role in the persister state. Under normal growth conditions, antitoxins potently inhibit the activities of the toxins. In contrast, under conditions of stress, the antitoxins are selectively degraded, freeing the toxins to inhibit essential cellular processes, such as DNA replication and protein translation. This inhibition results in rapid growth arrest. In this Review, we highlight recent discoveries of these multifaceted TA systems with a focus on the newly uncovered mechanisms, especially conditional cooperativity, that are used to regulate cell growth and persistence. We also discuss the potential for targeting TA systems for antimicrobial drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Neushul, P. Science, government, and the mass production of penicillin. J. Hist. Med. Allied Sci. 48, 371–395 (1993).
Balaban, N.Q. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775 (2011).
Bigger, J. Treatment of Staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Spoering, A.L. & Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183, 6746–6751 (2001).
Harrison, J.J. et al. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53, 2253–2258 (2009).
Keren, I., Shah, D., Spoering, A., Kaldalu, N. & Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Gerdes, K., Christensen, S.K. & Løbner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 (2005).
Magnuson, R.D. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189, 6089–6092 (2007).
Ogura, T. & Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 80, 4784–4788 (1983).
Gerdes, K., Rasmussen, P.B. & Molin, S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83, 3116–3120 (1986).
Lehnherr, H., Maguin, E., Jafri, S. & Yarmolinsky, M.B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233, 414–428 (1993).
Gotfredsen, M. & Gerdes, K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29, 1065–1076 (1998).
Hayes, F. & Van Melderen, L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408 (2011).
Christensen, S.K. et al. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51, 1705–1717 (2004).
Van Melderen, L., Bernard, P. & Couturier, M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11, 1151–1157 (1994).
Ramage, H.R., Connolly, L.E. & Cox, J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767 (2009).
Balaban, N.Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Kussell, E., Kishony, R., Balaban, N.Q. & Leibler, S. Bacterial persistence: a model of survival in changing environments. Genetics 169, 1807–1814 (2005).
Thisted, T., Sørensen, N.S., Wagner, E.G. & Gerdes, K. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13, 1960–1968 (1994).
Gerdes, K., Nielsen, A., Thorsted, P. & Wagner, E.G. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 226, 637–649 (1992).
Brantl, S. & Jahn, N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 39, 413–427 (2015).
Pedersen, K. & Gerdes, K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32, 1090–1102 (1999).
Pandey, D.P. & Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 (2005).
Afif, H., Allali, N., Couturier, M. & Van Melderen, L. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 41, 73–82 (2001).
Bernard, P. et al. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 234, 534–541 (1993).
Brown, B.L., Lord, D.M., Grigoriu, S., Peti, W. & Page, R. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J. Biol. Chem. 288, 1286–1294 (2013).
Wang, X. et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366 (2011).
Jiang, Y., Pogliano, J., Helinski, D.R. & Konieczny, I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979 (2002).
Christensen-Dalsgaard, M., Jørgensen, M.G. & Gerdes, K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348 (2010).
Pedersen, K. et al. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 (2003).
Zhang, Y. et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923 (2003).
Yamaguchi, Y., Park, J.H. & Inouye, M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284, 28746–28753 (2009).
Castro-Roa, D. et al. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 (2013).
Cruz, J.W. et al. Doc toxin is a kinase that inactivates elongation factor Tu. J. Biol. Chem. 289, 7788–7798 (2014).
Germain, E., Castro-Roa, D., Zenkin, N. & Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254 (2013).
Kaspy, I. et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4, 3001 (2013).
Brown, B.L. et al. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5, e1000706 (2009).
Kamada, K., Hanaoka, F. & Burley, S.K. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11, 875–884 (2003).
Neubauer, C. et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 (2009).
Christensen, S.K. & Gerdes, K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48, 1389–1400 (2003).
Feng, S. et al. YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 41, 9549–9556 (2013).
Zhang, Y., Yamaguchi, Y. & Inouye, M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 284, 25522–25531 (2009).
Maehigashi, T., Ruangprasert, A., Miles, S.J. & Dunham, C.M. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res. 43, 8002–8012 (2015).
Hurley, J.M. & Woychik, N.A. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613 (2009).
Schureck, M.A. et al. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J. Biol. Chem. 289, 1060–1070 (2014).
Schureck, M.A., Dunkle, J.A., Maehigashi, T., Miles, S.J. & Dunham, C.M. Defining the mRNA recognition signature of a bacterial toxin protein. Proc. Natl. Acad. Sci. USA 112, 13862–13867 (2015).
Christensen, S.K., Pedersen, K., Hansen, F.G. & Gerdes, K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819 (2003).
Arbing, M.A. et al. Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems. Structure 18, 996–1010 (2010).
Bøggild, A. et al. The crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity. Structure 20, 1641–1648 (2012).
Schumacher, M.A. et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401 (2009).
Fineran, P.C. et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 106, 894–899 (2009).
Short, F.L. et al. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc. Natl. Acad. Sci. USA 110, E241–E249 (2013).
Brown, J.M. & Shaw, K.J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185, 6600–6608 (2003).
Masuda, H., Tan, Q., Awano, N., Wu, K.P. & Inouye, M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84, 979–989 (2012).
Wang, X. et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861 (2012).
Wang, X. et al. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 15, 1734–1744 (2013).
Aakre, C.D., Phung, T.N., Huang, D. & Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628 (2013).
Moyed, H.S. & Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775 (1983).
Kim, Y. & Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391, 209–213 (2010).
Maisonneuve, E., Shakespeare, L.J., Jørgensen, M.G. & Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 108, 13206–13211 (2011).
Schumacher, M.A. et al. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524, 59–64 (2015).
Germain, E., Roghanian, M., Gerdes, K. & Maisonneuve, E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. USA 112, 5171–5176 (2015).
Helaine, S. et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014).
Verstraeten, N. et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 (2015).
Rotem, E. et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 107, 12541–12546 (2010).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013).
Garcia-Pino, A. et al. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142, 101–111 (2010).
Overgaard, M., Borch, J., Jørgensen, M.G. & Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69, 841–857 (2008).
Guglielmini, J. & Van Melderen, L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob. Genet. Elements 1, 283–290 (2011).
Dienemann, C., Bøggild, A., Winther, K.S., Gerdes, K. & Brodersen, D.E. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J. Mol. Biol. 414, 713–722 (2011).
Garcia-Pino, A. et al. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 283, 30821–30827 (2008).
De Jonge, N. et al. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell 35, 154–163 (2009).
Choy, M.S. et al. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci. USA 111, 4097–4102 (2014).
Peti, W., Nairn, A.C. & Page, R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 280, 596–611 (2013).
Wright, P.E. & Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015).
Gough, J. & Dunker, A.K. Sequences and topology: disorder, modularity, and post/pre translation modification. Curr. Opin. Struct. Biol. 23, 417–419 (2013).
Oberer, M., Zangger, K., Gruber, K. & Keller, W. The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding. Protein Sci. 16, 1676–1688 (2007).
Madl, T. et al. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J. Mol. Biol. 364, 170–185 (2006).
van der Lee, R. et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 8, 1832–1844 (2014).
Ragusa, M.J. et al. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 17, 459–464 (2010).
Loris, R. & Garcia-Pino, A. Disorder- and dynamics-based regulatory mechanisms in toxin-antitoxin modules. Chem. Rev. 114, 6933–6947 (2014).
Magnuson, R. & Yarmolinsky, M.B. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180, 6342–6351 (1998).
Johnson, E.P., Strom, A.R. & Helinski, D.R. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J. Bacteriol. 178, 1420–1429 (1996).
Monti, M.C. et al. Interactions of Kid-Kis toxin-antitoxin complexes with the parD operator-promoter region of plasmid R1 are piloted by the Kis antitoxin and tuned by the stoichiometry of Kid-Kis oligomers. Nucleic Acids Res. 35, 1737–1749 (2007).
Cataudella, I., Sneppen, K., Gerdes, K. & Mitarai, N. Conditional cooperativity of toxin-antitoxin regulation can mediate bistability between growth and dormancy. PLoS Comput. Biol. 9, e1003174 (2013).
Gelens, L., Hill, L., Vandervelde, A., Danckaert, J. & Loris, R. A general model for toxin-antitoxin module dynamics can explain persister cell formation in E. coli. PLoS Comput. Biol. 9, e1003190 (2013).
Ruangprasert, A. et al. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J. Biol. Chem. 289, 20559–20569 (2014).
Brown, B.L., Wood, T.K., Peti, W. & Page, R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 286, 2285–2296 (2011).
Centers for Disease Control and Prevention. in Antibiotic/Antimicrobial Resistance Vol. http://www.cdc.gov/drugresistance/ (2015).
Conlon, B.P. et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370 (2013).
Barbosa, L.C. et al. Design and synthesis of peptides from bacterial ParE toxin as inhibitors of topoisomerases. Eur. J. Med. Chem. 54, 591–596 (2012).
Kamada, K. & Hanaoka, F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19, 497–509 (2005).
Simanshu, D.K., Yamaguchi, Y., Park, J.H., Inouye, M. & Patel, D.J. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 52, 447–458 (2013).
Butt, A. et al. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem. J. 459, 333–344 (2014).
Brown, B.L. & Page, R. Preliminary crystallographic analysis of the Escherichia coli antitoxin MqsA (YgiT/b3021) in complex with mqsRA promoter DNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1060–1063 (2010).
Hargreaves, D. et al. Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure 10, 1425–1433 (2002).
Rao, F. et al. Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system. Nucleic Acids Res. 43, 9529–9540 (2015).
Acknowledgements
The authors thank all current and previous members of the Page and Peti laboratories for their contributions to this ongoing research. The Peti and Page laboratories also thank all collaborators in the toxin-antitoxin field for their critical scientific input and generous support, especially T.K. Wood (Pennsylvania State University). Grant sponsors: US National Science Foundation, grant number MCB0952550, and a Brown University Deans Award to R.P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Page, R., Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 12, 208–214 (2016). https://doi.org/10.1038/nchembio.2044
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2044
This article is cited by
-
MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding
Nature Communications (2023)
-
Role of PumB antitoxin as a transcriptional regulator of the PumAB type-II toxin–antitoxin system and its endoribonuclease activity on the PumA (toxin) transcript
Molecular Genetics and Genomics (2023)
-
Membrane Proteins as a Regulator for Antibiotic Persistence in Gram-Negative Bacteria
Journal of Microbiology (2023)
-
VapC toxin switches M. smegmatis cells into dormancy through 23S rRNA cleavage
Archives of Microbiology (2023)
-
High-resolution genomic surveillance elucidates a multilayered hierarchical transfer of resistance between WWTP- and human/animal-associated bacteria
Microbiome (2022)