Abstract
P2X receptors are trimeric, non-selective cation channels activated by ATP that have important roles in the cardiovascular, neuronal and immune systems. Despite their central function in human physiology and although they are potential targets of therapeutic agents, there are no structures of human P2X receptors. The mechanisms of receptor desensitization and ion permeation, principles of antagonism, and complete structures of the pore-forming transmembrane domains of these receptors remain unclear. Here we report X-ray crystal structures of the human P2X3 receptor in apo/resting, agonist-bound/open-pore, agonist-bound/closed-pore/desensitized and antagonist-bound/closed states. The open state structure harbours an intracellular motif we term the ‘cytoplasmic cap’, which stabilizes the open state of the ion channel pore and creates lateral, phospholipid-lined cytoplasmic fenestrations for water and ion egress. The competitive antagonists TNP-ATP and A-317491 stabilize the apo/resting state and reveal the interactions responsible for competitive inhibition. These structures illuminate the conformational rearrangements that underlie P2X receptor gating and provide a foundation for the development of new pharmacological agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Londos, C., Cooper, D. M. & Wolff, J. Subclasses of external adenosine receptors. Proc. Natl Acad. Sci. USA 77, 2551–2554 (1980)
van Calker, D., Müller, M. & Hamprecht, B. Adenosine regulates via two different types of receptors, the accumulation of cyclic AMP in cultured brain cells. J. Neurochem. 33, 999–1005 (1979)
Burnstock, G. Do some nerve cells release more than one transmitter? Neuroscience 1, 239–248 (1976)
Valera, S. et al. A new class of ligand-gated ion channel defined by P2x receptor for extracellular ATP. Nature 371, 516–519 (1994)
Webb, T. E. et al. Cloning and functional expression of a brain G-protein-coupled ATP receptor. FEBS Lett. 324, 219–225 (1993)
Fountain, S. J. & Burnstock, G. An evolutionary history of P2X receptors. Purinergic Signal. 5, 269–272 (2009)
Burnstock, G. & Kennedy, C. P2X receptors in health and disease. Adv. Pharmacol. 61, 333–372 (2011)
Surprenant, A. & North, R. A. Signaling at purinergic P2X receptors. Annu. Rev. Physiol. 71, 333–359 (2009)
North, R. A. & Jarvis, M. F. P2X receptors as drug targets. Mol. Pharmacol. 83, 759–769 (2013)
Brake, A. J., Wagenbach, M. J. & Julius, D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature 371, 519–523 (1994)
North, R. A. Molecular physiology of P2X receptors. Physiol. Rev. 82, 1013–1067 (2002)
Habermacher, C., Dunning, K., Chataigneau, T. & Grutter, T. Molecular structure and function of P2X receptors. Neuropharmacology 104, 18–30 (2016)
Egan, T. M. & Khakh, B. S. Contribution of calcium ions to P2X channel responses. J. Neurosci. 24, 3413–3420 (2004)
Virginio, C., Robertson, G., Surprenant, A. & North, R. A. Trinitrophenyl-substituted nucleotides are potent antagonists selective for P2X1, P2X3, and heteromeric P2X2/3 receptors. Mol. Pharmacol. 53, 969–973 (1998)
Jarvis, M. F. & Khakh, B. S. ATP-gated P2X cation-channels. Neuropharmacology 56, 208–215 (2009)
Koshimizu, T., Koshimizu, M. & Stojilkovic, S. S. Contributions of the C-terminal domain to the control of P2X receptor desensitization. J. Biol. Chem. 274, 37651–37657 (1999)
Allsopp, R. C. & Evans, R. J. The intracellular amino terminus plays a dominant role in desensitization of ATP-gated P2X receptor ion channels. J. Biol. Chem. 286, 44691–44701 (2011)
Allsopp, R. C., Farmer, L. K., Fryatt, A. G. & Evans, R. J. P2X receptor chimeras highlight roles of the amino terminus to partial agonist efficacy, the carboxyl terminus to recovery from desensitization, and independent regulation of channel transitions. J. Biol. Chem. 288, 21412–21421 (2013)
Boué-Grabot, E., Archambault, V. & Séguéla, P. A protein kinase C site highly conserved in P2X subunits controls the desensitization kinetics of P2X(2) ATP-gated channels. J. Biol. Chem. 275, 10190–10195 (2000)
Brändle, U. et al. Desensitization of the P2X(2) receptor controlled by alternative splicing. FEBS Lett. 404, 294–298 (1997)
Hausmann, R. et al. A hydrophobic residue in position 15 of the rP2X3 receptor slows desensitization and reveals properties beneficial for pharmacological analysis and high-throughput screening. Neuropharmacology 79, 603–615 (2014)
Koshimizu, T., Tomic´, M., Koshimizu, M. & Stojilkovic, S. S. Identification of amino acid residues contributing to desensitization of the P2X2 receptor channel. J. Biol. Chem. 273, 12853–12857 (1998)
Smith, F. M., Humphrey, P. P. & Murrell-Lagnado, R. D. Identification of amino acids within the P2X2 receptor C-terminus that regulate desensitization. J. Physiol. (Lond.) 520, 91–99 (1999)
Werner, P., Seward, E. P., Buell, G. N. & North, R. A. Domains of P2X receptors involved in desensitization. Proc. Natl Acad. Sci. USA 93, 15485–15490 (1996)
Zemkova, H., He, M. L., Koshimizu, T. A. & Stojilkovic, S. S. Identification of ectodomain regions contributing to gating, deactivation, and resensitization of purinergic P2X receptors. J. Neurosci. 24, 6968–6978 (2004)
Fryatt, A. G. & Evans, R. J. Kinetics of conformational changes revealed by voltage-clamp fluorometry give insight to desensitization at ATP-gated human P2X1 receptors. Mol. Pharmacol. 86, 707–715 (2014)
Hattori, M. & Gouaux, E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature 485, 207–212 (2012)
Kasuya, G. et al. Structural insights into divalent cation modulations of ATP-gated P2X receptor channels. Cell Reports 14, 932–944 (2016)
Kawate, T., Michel, J. C., Birdsong, W. T. & Gouaux, E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 460, 592–598 (2009)
Habermacher, C. et al. Photo-switchable tweezers illuminate pore-opening motions of an ATP-gated P2X ion channel. eLife 5, e11050 (2016)
Heymann, G. et al. Inter- and intrasubunit interactions between transmembrane helices in the open state of P2X receptor channels. Proc. Natl Acad. Sci. USA 110, E4045–E4054 (2013)
Zhou, H. X. & Cross, T. A. Influences of membrane mimetic environments on membrane protein structures. Annu. Rev. Biophys. 42, 361–392 (2013)
Minato, Y. et al. Conductance of P2X4 purinergic receptor is determined by conformational equilibrium in the transmembrane region. Proc. Natl Acad. Sci. USA 113, 4741–4746 (2016)
Quick, M. & Javitch, J. A. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl Acad. Sci. USA 104, 3603–3608 (2007)
Chen, C. C. et al. A P2X purinoceptor expressed by a subset of sensory neurons. Nature 377, 428–431 (1995)
Lewis, C. et al. Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature 377, 432–435 (1995)
Burgard, E. C. et al. Competitive antagonism of recombinant P2X(2/3) receptors by 2′, 3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate (TNP-ATP). Mol. Pharmacol. 58, 1502–1510 (2000)
Jarvis, M. F. et al. A-317491, a novel potent and selective non-nucleotide antagonist of P2X3 and P2X2/3 receptors, reduces chronic inflammatory and neuropathic pain in the rat. Proc. Natl Acad. Sci. USA 99, 17179–17184 (2002)
Egan, T. M., Haines, W. R. & Voigt, M. M. A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J. Neurosci. 18, 2350–2359 (1998)
Hille, B. Ion Channels of Excitable Membranes 3rd edn (Sinauer, 2001)
Degrève, L., Vechi, S. M. & Junior, C. Q. The hydration structure of the Na+ and K+ ions and the selectivity of their ionic channels. Biochim. Biophys. Acta 1274, 149–156 (1996)
Migita, K., Haines, W. R., Voigt, M. M. & Egan, T. M. Polar residues of the second transmembrane domain influence cation permeability of the ATP-gated P2X(2) receptor. J. Biol. Chem. 276, 30934–30941 (2001)
Jiang, R. et al. Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J. 31, 2134–2143 (2012)
Li, M., Kawate, T., Silberberg, S. D. & Swartz, K. J. Pore-opening mechanism in trimeric P2X receptor channels. Nat. Commun. 1, 44 (2010)
Fujiwara, Y., Keceli, B., Nakajo, K. & Kubo, Y. Voltage- and [ATP]-dependent gating of the P2X(2) ATP receptor channel. J. Gen. Physiol. 133, 93–109 (2009)
Khakh, B. S., Bao, X. R., Labarca, C. & Lester, H. A. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat. Neurosci. 2, 322–330 (1999)
Li, M., Silberberg, S. D. & Swartz, K. J. Subtype-specific control of P2X receptor channel signaling by ATP and Mg2+. Proc. Natl Acad. Sci. USA 110, E3455–E3463 (2013)
Giniatullin, R., Sokolova, E. & Nistri, A. Modulation of P2X3 receptors by Mg2+ on rat DRG neurons in culture. Neuropharmacology 44, 132–140 (2003)
Kawate, T., Robertson, J. L., Li, M., Silberberg, S. D. & Swartz, K. J. Ion access pathway to the transmembrane pore in P2X receptor channels. J. Gen. Physiol. 137, 579–590 (2011)
Samways, D. S., Khakh, B. S., Dutertre, S. & Egan, T. M. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors). Proc. Natl Acad. Sci. USA 108, 13800–13805 (2011)
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014)
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010)
Hanson, M. A. et al. Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010)
Li, H., Robertson, A. D. & Jensen, J. H. Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 (2005)
Olsson, M. H., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011)
Søndergaard, C. R., Olsson, M. H., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011)
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998)
Mackerell, A. D., Jr, Feig, M. & Brooks, C. L., III . Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 (2004)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994)
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995)
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993)
Acknowledgements
We thank M. Hattori for initial construct screening, L. Vaskalis for figures, H. Owen for manuscript preparation and Gouaux laboratory members for discussions. We acknowledge the Berkeley Center for Structural Biology at the Advanced Light source for assistance with data collection at beamline 5.0.2 and the Northeastern Collaborative Access Team at the Advanced Photon Source for assistance with data collection at beamline 24-ID-C. The simulations were supported by the National Institutes of General Medical Sciences (U54-GM087519 and P41-GM104601 to E.T.) and computationally through XSEDE (TG-MCA06N060 to E.T.). E.G. is an investigator with the Howard Hughes Medical Institute. This research was supported by the National Institute of General Medical Sciences (5F32GM108391 to S.E.M and R01GM100400 to E.G.).
Author information
Authors and Affiliations
Contributions
S.E.M. and E.G. designed the project. S.E.M. performed the biochemical and functional analyses. S.E.M. and W.O. carried out the protein purification and crystallization. S.E.M., W.L., and W.O. performed the crystallography and model building. M.S. and E.T. performed the molecular dynamics simulations. All authors wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks P. Biggin, R. Murrell-Lagnado and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
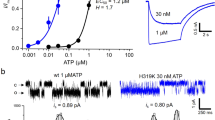
Extended Data Figure 1 Functional studies of hP2X3-MFC and hP2X3-MFCslow.
a, Measurement of [3H-ATP] saturation binding to purified, detergent-solubilized hP2X3-MFC using SPA. For each point in the plot, the error bars indicate the standard error of the mean (SEM) for triplicate samples. The calculated Kd for ATP binding was 2.8 ± 0.1 nM and represents the average of two separate experiments. b, ATP-induced currents for hP2X3-WT and hP2X3-MFC both show rapid desensitization kinetics with τ = 523 ± 198 ms and 429 ± 43 ms, respectively. These values represent an average of three measurements with error values indicating s.e.m. Actual rate constants are likely to be faster as the perfusion rate of our TEVC system is ~1,000 ms. c, Measurement of [3H-ATP] saturation binding to purified, detergent solubilized hP2X3-MFCslow using SPA. The calculated Kd for ATP binding was 3.3 ± 0.3 nM. d, ATP-induced currents for hP2X3-MFCslow show delayed desensitization kinetics with τ = 42,581 ± 2,194 ms. e, f, Co-application of 2 μM TNP-ATP (e) or 2 μM A-317491 (f) inhibits the current induced by 1 μM ATP for hP2X3-WT, hP2X3-MFC and hP2X3-MFCslow. g, Inhibition of 3H-ATP binding to hP2X3-MFCslow by unlabelled TNP-ATP yields a Ki of 94 ± 12 nM. Inhibition of 3H-ATP binding to hP2X3-MFC by unlabelled TNP-ATP yields a Ki of 118 ± 1 nM (data not shown). For each point in the plot, the error bars indicate the s.e.m. for triplicate samples. The reported Ki values represent the average of two separate experiments. h, i, Co-application of 2 μΜ TNP-ATP (h) or 2 μΜ A-317491 (i) blocks the residual current remaining after prolonged application of 1 μM ATP to hP2X3-MFCslow receptors.
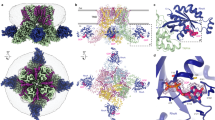
Extended Data Figure 2 Naming of purinergic receptor domains and comparison of hP2X3 structures to previously published zfP2X4 structures.
a, Ribbon representation of one subunit of the open state structure of hP2X3 receptor shown in orthogonal views. The new cytoplasmic cap domain is termed the ‘tail fin’. b, Cartoon representation of the open state hP2X3 structure aligned to the open state zfP2X4 structure (construct name ΔP2X4-C) shown parallel to the membrane as a side view and as viewed perpendicular to the membrane from the extracellular side. The transmembrane domains for the hP2X3 structure are longer and more complete than for the zfP2X4 structure. c, Cartoon representation of the apo state hP2X3 structure aligned to the apo state zfP2X4 structure (construct name ΔP2X4-B) shown parallel to the membrane as a side view and viewed perpendicular to the membrane from the extracellular side. d, Sequence alignment of the N terminus (top alignment) and C terminus (bottom alignment) of hP2X3 compared to zfP2X4. Starting and ending residues of the hP2X3 construct compared to the ΔP2X4-C construct are indicated with red arrows. The hP2X3 crystallization construct has more residues at both termini than the ΔP2X4-C crystallization construct.
Extended Data Figure 3 The pore-lining surface of hP2X3 for the open, apo and desensitized states.
a, A coronal section of a surface representation of the open state of hP2X3 reveals that four vestibules (upper, central, extracellular and intracellular) are located on the molecular three-fold axis. b, Pore-lining surfaces along the entire axis of hP2X3 for open, apo and desensitized states. The colour of each sphere represents a different radius from the receptor centre, as calculated by the program HOLE: red <1.15 Å, green 1.15–2.30 Å, purple >2.30 Å. c, Plot of pore radius as a function of distance along the pore axis for the open state versus the apo state versus the desensitized state. The positions of the residues making up the narrowest radius in each conformational state are labelled. The Cα position of I341 is set as zero. I323 defines the first constriction site of the gate (extracellular boundary of the gate), whereas T330 defines the second constriction site (cytoplasmic boundary of the gate). These residues are at the equivalent positions that define the boundaries of the gate in the apo state structure of zfP2X4, but are leucine and alanine residues, respectively, in zfP2X4. A single residue, V334, defines the constriction site of the desensitized state. Residue T330 defines the narrowest region of the pore in the open state.
Extended Data Figure 4 The overall structure and ion channel pore of antagonist-bound/closed states.
a, b, Cartoon representation of the competitive antagonist-bound/closed state structures, TNP-ATP in cyan (a) and A-317491 in blue (b), shown parallel to the membrane as a side view. c, An overall alignment of a single protomer in the apo state (red-purple), TNP-ATP-bound state (cyan) and A-317491-bound state (blue). d, Plot of pore radius as a function of distance along the pore axis for apo state versus TNP-ATP-bound state versus A-317491-bound state. The positions of the residues making up the narrowest radius in each conformational state are labelled. The Cα position of I341 is set as zero. e, f, Pore-lining surfaces along the entire axis of the receptor and a focus on the transmembrane domain with TM2 pore-lining residues shown as sticks for the TNP-ATP-bound state (e) and the A-317491-bound state (f). The colour of each sphere represents a different radius from the receptor centre, as calculated by the program HOLE: red <1.15 Å, green 1.15–2.30 Å, purple >2.30 Å.
Extended Data Figure 5 High-affinity P2X3 agonist 2-methylthio-ATP can be soaked into the desensitized state crystals.
a, Inhibition of 3H-ATP binding to hP2X3-MFC by unlabelled 2-methylthio-ATP yields a Ki of 1.9 ± 0.1 nM. For each point in the plot, the error bars indicate the s.e.m. for triplicate samples. The reported Ki represents the average of two separate experiments. b, Electron density for ATP in the desensitized state. The Fo−Fc map is contoured at 1.0σ. c, Desensitized state crystals that have been soaked with 2-methylthio-ATP have a density in the binding pocket, which matches the shape of 2-methylthio-ATP, accounting for the methyl-thio moiety. The Fo−Fc map is contoured at 1.0σ. d, An anomalous difference Fourier map (contoured at 3.0σ) has anomalous signal that overlaps with the sulfur moiety of 2-methylthio-ATP as well as the phosphate groups. These crystals of hP2X3-MFC successfully ligand-exchanged ATP for agonist 2-methylthio-ATP in the binding pocket but were destroyed when soaked with antagonist TNP-ATP, providing evidence that the structure represents an agonist-bound, closed or desensitized state.
Extended Data Figure 6 Resetting from desensitized to apo state of hP2X3.
a, b, Structures of hP2X3 in the desensitized state (a) and apo state (b) shown parallel to the membrane. There are marked changes between the two states in the extracellular domain and the transmembrane domain. c, d, Top-down view comparing the pore of the desensitized state (c) to the pore of the apo state (d) highlighting how, although both pores are closed, the residues that define the gates are different. e, Relative differences in the pore between desensitized and apo states after aligning the upper body domain of the trimer reveal that a significant clockwise conformational change at both the extracellular and cytoplasmic surfaces of the transmembrane domain must occur for the receptor pore to reset back to the apo state. f, Alignment of TM2 in desensitized versus apo state shows that both helices have the same helical pitch, suggesting that the 310-helix that existed in the open state is a transient structural feature. The inset shows the view along the axis of the TM2 helix, observed from the cytoplasmic surface. We speculate that the structural resetting of the receptor from the desensitized state to the apo state is likely to occur after ligand dissociation.
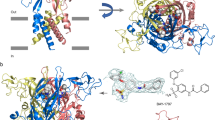
Extended Data Figure 7 Orthosteric ligand-binding site and ligand densities.
a, b, View of the orthosteric binding pocket for the ATP-bound open state structure of hP2X3. ATP binds at an interface between two protomers, with protomer A shown in green and protomer B shown in grey. The 2Fo−Fc density for ATP is shown at 2.5σ. c, d, View of the orthosteric binding pocket for the TNP-ATP-bound closed state structure of hP2X3 with protomer A shown in cyan and protomer B shown in grey. The 2 Fo−Fc density for TNP-ATP is shown at 1.5σ. e, f, View of the orthosteric binding pocket for the A-317491-bound closed state structure of hP2X3 with protomer A shown in blue and protomer B shown in grey. The 2Fo−Fc density for A-317491 is shown at 0.8σ. g, Close-up comparison of the relative orientation of ATP (shown as translucent) versus TNP-ATP in the binding pocket highlights how the phosphate moiety and the orientation of the ribose group are both inverted between the two molecules. h, The apo state structure (shown in figure) as well as both antagonist-bound structures have a Mg2+ ion present in the head domain of hP2X3, coordinated by the side chains of E109 and D158 as well as the carbonyl oxygen of E156. The 2Fo−Fc density for the Mg2+ ion is shown at 1.5σ.
Extended Data Figure 8 Anomalous signal from Mn2+ ion proves Mg2+ ion is present in the head domain of the apo state.
a, Anomalous difference map of apo structure with crystals grown in MnCl2 have an anomalous signal from a Mn2+ ion in the head domain (anomalous difference Fourier map shown in green contoured at 5.5σ). This anomalous signal from Mn2+ overlaps with the 2Fo−Fc density shown in Extended Data Fig. 7h, proving this density is a Mg2+ ion. b, The Mn2+ ion in the head domain is coordinated by the side chains of E109 and D158 and the carbonyl oxygen of E156. c, The presence of a Mg2+ ion does not change the affinity of ATP for hP2X3-MFCslow, as assessed by SPA binding, suggesting that Mg2+ does not compete with ATP for the binding pocket or impair the ability of ATP to bind to the receptor. For each point in the plot, the error bars indicate the s.e.m. for triplicate measurements. The reported Kd values represent the mean of two separate experiments.
Supplementary information
Demonstration of the overall structural conformational changes that occur during the gating cycle of hP2X3 receptor
This video demonstrates the overall structural conformational changes that occur during the gating cycle of hP2X3 receptor, highlighting the receptor's transition from the apo state to the open state to the desensitized state before resetting back to the apo state. Each protomer subunit is shown in a different colour. (MOV 9747 kb)
Structural conformational changes that occur in the receptor's pore during the gating cycle of hP2X3, highlighting the transition of the pore from the apo state to the open state to the desensitized state before resetting back to the apo state.
This video demonstrates the structural conformational changes that occur in the receptor's pore during the gating cycle of hP2X3, shown perpendicular to the membrane from the extracellular surface. It highlights the transition of the pore from the apo state to the open state to the desensitized state before resetting back to the apo state. Each protomer subunit is shown in a different colour. (MOV 10426 kb)
Structural conformational changes that occur in the receptor’s pore during the gating cycle of hP2X3, highlighting the residues that define the narrowest region across the pore in each conformational state: I323 in the apo state (red-purple), T330 in the open state (green) and V334 in the desensitized state (yellow).
This video shows the structural conformational changes that occur in the receptor’s pore during the gating cycle of hP2X3, shown perpendicular to the membrane from the extracellular surface, highlighting the residues that define the narrowest region across the pore in each conformational state: I323 in the apo state (red-purple), T330 in the open state (green), and V334 in the desensitized state (yellow). The movie steps through the transition of the pore from the apo state to the open state to the desensitized state before resetting back to the apo state. (MOV 9739 kb)
Rights and permissions
About this article
Cite this article
Mansoor, S., Lü, W., Oosterheert, W. et al. X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature 538, 66–71 (2016). https://doi.org/10.1038/nature19367
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19367
This article is cited by
-
Role of ATP in migraine mechanisms: focus on P2X3 receptors
The Journal of Headache and Pain (2023)
-
Optical control of PIEZO1 channels
Nature Communications (2023)
-
Chronic cough relief by allosteric modulation of P2X3 without taste disturbance
Nature Communications (2023)
-
Triggering of Major Brain Disorders by Protons and ATP: The Role of ASICs and P2X Receptors
Neuroscience Bulletin (2023)
-
DT-0111: a novel P2X3 receptor antagonist
Purinergic Signalling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.