Abstract
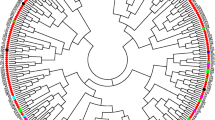
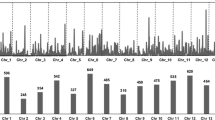
A series of 320 mapped simple sequence repeats (SSRs) have been used to screen the allelic diversity of tetraploid Gossypium species. Fourty-seven genotypes were analyzed representing (i) the wide spectrum of diversity of the cultivated pool and of the primitive landraces of species G. hirsutum (‘marie-galante’, ‘punctatum’, ‘richmondi’, ‘morrilli’, ‘palmeri’, and ‘latifolium’, and ‘yucatanense’), and (ii) species G. barbadense, G. darwinii and G. tomentosum. The polymorphism of 201 SSR loci revealed 1128 allelic variants ranging from 3 to 17 per locus. Neighbor-joining (NJ) method based on genetic dissimilarities produced groupings consistent with the assignments of accessions both at species and at race level. Our data confirmed the proximity of the Galapagos endemic species G. darwinii to species G. barbadense. Within species G. hirsutum, and as compared to the other 6 races, race yucatanense appeared as the most distant from cultivated genotypes. Race yucatanense also exhibited the highest number of unique alleles. The important informative heterogeneity of the 201 SSR loci was exploited to select the most polymorphic ones that were assembled into three series of genome-wide (i.e. each homoeologous AD chromosome pair being equally represented) and mutliplexable (× 3) SSRs. Using one of these ‘genotyping set’, consisting of 39 SSRs (one 3-plex for each of the 13 AD chromosomes pairs) or 45 loci, we were able to assess the relationships between accessions and the topology in the genetic diversity sampled. Such genotyping set of highly informative SSR markers assembled in PCR-multiplex, while increasing genotyping throughput, will be applicable for molecular genetic diversity studies of large germplasm collections.



Similar content being viewed by others
References
Abdalla AM, Reddy OUK, El-Zik KM, Pepper AE (2001) Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet 102:222–229
Adams KL, Wendel JF (2004) Exploring the mysteries of polyploidy in cotton. Biol J Linn Soc 82:573–581
Ahoton L, Lacape J-M, Baudoin J-P, Mergeai G (2003) Introduction of australian diploid cotton genetic variation into upland cotton. Crop Sci 43:1999–2005
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Ano G, Schwendiman J, Fersing J, Lacape J-M (1982) Les cotonniers primitifs de G. hirsutum race yucatanense de la Pointe des Châteaux en Guadeloupe et l’origine possible des cotonniers tétraploides du Nouveau Monde. Cot Fib Trop 37(4):327–332
Bourdon C (1984) Différenciation génétique inter et intraspécifique dans le genre Gossypium L. : le polymorphisme enzymatique chez des espèces diploïdes et tétraploïdes de cotonnier, PhD Dissertation, Université Paris Sud, Paris, pp. 172
Brubaker CL, Wendel JF (1993) On the specific status of Gossypium lanceolatum Todaro. Genet Res Crop Evol 40:165–170
Brubaker CL, Wendel JF (1994) Reevaluating the origin of domesticated cotton (Gossypium hirsutum, Malvaceae) using nuclear restriction fragment length polymorphism (RFLP). Am J Bot 81:1309–1326
Brubaker CL, Bourland FM, Wendel JF (1999) The origin and domestication of cotton. In: Smith CW, Cothren JT (eds) Cotton. origin, history, technology, and production. John Wiley & Sons, New York, pp 3–31
Coburn JR, Temnykh S, Paul EM, McCouch S (2002) Design and application of microsatellite marker panels for semi-automated genotyping of rice (Oryza sativa L). Crop Sci 42:2092–2099
Dejoode DR, Wendel JF (1992) Genetic diversity and origin of the Hawaiian islands cotton, Gossypium tomentosum. Am J Bot 79:1311–1319
Diwan N, Cregan PB (1997) Automated sizing of fluorescent-labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor Appl Genet 95:723–733
Ellegren H (2000) Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet 16:551–558
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Han ZG, Guo W, Song XL, Zhang T (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Genomics 272:308–327
Han ZG, Wang C, Song XL, Guo W, Gou JY, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Hutchinson JB (1951) Intra-specific differentiation in Gossypium hirsutum. Heredity 5:169–193
Iqbal J, Reddy OUK, El-Zik KM, Pepper AE (2001) A genetic bottleneck in the ‘evolution under domestication’ of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor Appl Genet 103:547–554
Jain S, Jain RK, McCouch S (2004) Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor Appl Genet 109:965–977
Khan SA, Hussain D, Askari E, Stewart JM, Malik KA, Zafar Y (2000) Molecular phylogeny of Gossypium species by DNA fingerprinting. Theor Appl Genet 101:931–938
Lacape J-M, Nguyen TB, Thibivilliers S, Courtois B, Bojinov BM, Cantrell RG, Burr B, Hau B (2003) A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 46:612–626
Liu S, Cantrell RG, McCarty JCJ, Stewart JM (2000) Simple sequence repeat-based assessment of genetic diversity in cotton race stock accessions. Crop Sci 40:1459–1469
Liu D, Guo XP, Lin Z, Nie YC, Zhang X (2005) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Res Crop Evol DOI: 10.1007/s10722-005-1304-y
Macaulay M, Ramsay L, Powell W, Waugh R (2001) A representative, highly informative ‘genotyping set’ of barley SSRs. Theor Appl Genet 106:801–809
Maccaferri M, Sanguineti MC, Donini P, Tuberosa R (2003) Microsatellite analysis reveals a progressive widening of the genetic basis in the elite durum wheat germplasm. Theor Appl Genet 107(5):783–797
Masi P, Spagnoletti Zeuli PL, Donini P (2003) Development and analysis of multiplex microsatellite markers sets in common bean (Phaseolus vulgaris L). Mol Breed 11:303–313
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nguyen TB, Giband M, Brottier P, Risterucci A-M, Lacape J-M (2004) Wide coverage of tetraploid cotton genome using newly developed microsatellite markers. Theor Appl Genet 109:167–175
Park Y-H, Alabady MS, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, El-Shihy OM, Cantrell RG, Ulloa M (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line (RIL) cotton population. Mol Genet Genomics 274:428–441
Peakal R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Percival AE, Kohel RJ (1990) Distribution, collection, and evaluation of Gossypium. Adv Agronomy 44:225–256
Percival AE, Wendel JF, Stewart JM (1999) Taxonomy and germplasm resources. In: Smith CW, Cothren JT (eds) Cotton origin, history, technology, and production. John Wiley & Sons Inc., New York, pp 33–63
Perrier X, Flori A, Bonnot F (2003) Methods of data analysis. In: Hamon S, Seguin M, Perrier X, Glaszmann J-C (eds) Genetic diversity of cultivated tropical plants. CIRAD, Montpellier (France), pp 31–63
Pillay M, Myers GO (1999) Genetic diversity in cotton assessed by variation in ribosomal RNA genes and AFLP markers. Crop Sci 39:1881–1886
Ponce MR, Robles P, Micol JL (1999) High-throughput genetic mapping in Arabidopsis thaliana. Mol Gen Genet 261(2):408–415
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Powell W, Orozco-Castillo C, Chalmers KJ, Provan J, Waugh R (1995) Polymerase chain reaction-based assays for the characterization of plant genetic resources. Electrophoresis 16(9):1726–1730
Reddy OUK, Pepper AE, Abdurakhmonov I, Saha S, Jenkins JN, Brooks T, Bolek Y, El-Zik KM (2001) New dinucleotide and trinucleotide microsatellite marker resources for cotton genome research. J Cotton Sci 5:103–113
Reed PW, Davies JL, Copeman JB, Bennett ST, Palmer SM, Pritchard LE, Gouch SCL, Kawaguchi Y, Cordell HJ, Balfour KM, Jenkins SC, Powell EE, Vignal A, Todd JA (1994) Chromosome specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nature Genet 7:390–395
Rong J, Abbey C, Bowers JE, Brubaker CL, Chang C, Chee P, Delmonte TA, Ding X, Garza JJ, Marler BS, Park C-h, Pierce GJ, Rainey KM, Rastogi VK, Schulze SR, Trolinder N, Wendel JF, Wilkins TA, Williams-Coplin TD, Wing RA, Wright RJ, Zhao X, Zhu L, Paterson AH (2004) A 3347 locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166:389–417
Rungis D, Llewellyn D, Dennis ES, Lyon BR (2005) Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aus J Agr Res 56:301–307
Saghai Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW (1994) Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci USA 91(12):5466–5470
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Seignobos C, Schwendiman J (1991) Traditional cotton plants in Cameroon. Cot Fib Trop 46:322–332
Small RL, Ryburn JA, Wendel JF (1999) Low levels of nucleotide diversity at homoeologous Adh loci in allotetraploid cotton (Gossypium L.). Mol Biol Evol 16:491–501
Smulders MJM, Bredemeijer G, Rus-Kortekaas W, Arens P, Vosman B (1997) Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicon esculentum cultivars and accessions of other Lycopersicon species. Theor Appl Genet 97:264–272
Song QJ, Quigley CV, Nelson RL, Carter TE, Boerma HR, Strachan JL, Cregan PB (1999) A selected set of trinucleotide simple sequence repeat markers for soybean cultivar identification. Plant Var Seeds 12:207–220
Stewart JM (1994) Potential for crop improvement with exotic germplasm and genetic engineering. In: Constable GA, Forrester NW (eds) Challenging the future, Proceedings World Cotton Res. Conf. −1, Brisbane (AUS), pp 313–327
Struss D, Plieske J (1998) The use of microsatellite markers for detection of genetic diversity in barley populations. Theor Appl Genet 97:308–315
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Thuillet AC, Bataillon T, Sourdille P, David JL (2004) Factors affecting polymorphism at microsatellite loci in bread wheat [Triticum aestivum (L.) Thell]: effects of mutation processes and physical distance from the centromere. Theor Appl Genet 108(2):368–377
Tommasini L, Batley J, Arnold GM, Cooke RJ, Donini P, Lee D, Law JR, Lowe C, Moule C, Trick M, Edwards KJ (2003) The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106:1091–1101
Vigouroux Y, Jaqueth JS, Matsuoka Y, Smith OS, Beavis WD, Smith SC, Doebley J (2002) Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol 19:1251–1260
Wendel JF, Brubaker CL, Percival AE (1992) Genetic diversity in Gossypium hirsutum and the origin of Upland cotton. Am J Bot 79:1291–1310
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agronomy 78:140–186
Wendel JF, Percy RG (1990) Allozyme diversity and introgression in the Galapagos Islands endemic Gossypium darwinii and its relationship to continental G. barbadense. Biochem Syst Ecol 18:517–528
Acknowledgements
We thank Dr. Xavier Perrier for many helpful discussions and assistance with the statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lacape, JM., Dessauw, D., Rajab, M. et al. Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breeding 19, 45–58 (2007). https://doi.org/10.1007/s11032-006-9042-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-006-9042-1