Abstract
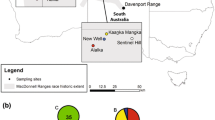
The eastern barred bandicoot, Perameles gunnii, has undergone a dramatic decline in distribution and abundance on the mainland of Australia during the twentieth century. In 1988 a captive breeding program was initiated to reduce the chance of extinction. With the extinction of the last wild mainland population in the early 1990s, reintroductions from captive-bred P. gunnii have met limited success, and currently only two extant populations persist in predator proof enclosures in the State of Victoria. With ~20 years of breeding, there are concerns that the genetic diversity within the breeding program has declined and may inhibit current and future success of the program. We have used ten nuclear microsatellite loci and sequencing of two partial mitochondrial genes (cytochrome oxidase I and ATPase 6) to determine genetic diversity within current Victorian P. gunnii. These diversity estimates are compared with historic samples from the captive breeding program dating back to 1995, historic samples from the last wild mainland population found at Hamilton in 1992 and contemporary Tasmanian wild populations. Results indicate that the captive P. gunnii population in the State of Victoria has lost significant genetic diversity through time. Genetic diversity is also reduced in populations at Hamilton Community Parklands and Mount Rothwell. Samples from the last wild population at Hamilton collected in 1992, along with samples from Tasmanian P. gunnii, had significantly greater genetic diversity than contemporary mainland populations. The results are discussed with reference to management options for maintaining genetic diversity within Victorian P. gunnii, including crossing Victorian and Tasmanian P. gunnii to increase genetic diversity, adaptability and evolutionary potential.





Similar content being viewed by others
References
Benjamini Y, Hochberg Y (2000) On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat 25:60–83
Blacket MJ, Robin C, Good RT, Miller AD (2012) Universal primers for fluorescent labeling of PCR fragments—an efficient and cost effective approach to genotyping by fluorescence. Mol Ecol Resour 12:456–463
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
Frankham R (1999) Quantitative genetics in conservation biology. Genet Res 74:237–244
Frankham R, Ballou JD, Eldridge MBD, Lacy RC et al (2011) Predicting the risk of outbreeding depression: critical information for managing fragmented populations. Conserv Biol 25:465–475
Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P (2010) Calculations of population differentiation based on G(ST) and D: forget G(ST) but not all of statistics! Mol Ecol 19:3845–3852
Goudet J (2001) FSTAT, a computer program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www.unil.ch/izea/popgen/softwares/fstat.htm
Hedrick PW (1995) Gene flow and genetic restoration: the Florida panther as a case study. Conserv Biol 5:996–1007
Hedrick PW (1999) Genetics of populations, 2nd edn. Jones and Bartlett Publishers, London
Hedrick PW, Fredrickson R (2010) Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv Genet 11:615–626
Hoffmann AA, Parsons PA (1997) Extreme environmental change and evolution. Cambridge University Press, Cambridge
Hood G (2002) PopTools. CSIRO, Canberra
Jost L (2008) G(ST) and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lewis PO, Zaykin D (2001) Genetic data analysis: computer program for the analysis of allelic data. http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php 20 August 2012
Lopez S, Rousset F, Shaw FH, Shaw RG, Ronce O (2009) Joint effects of inbreeding and local adaptation on the evolution of genetic load after fragmentation. Conserv Biol 23:1618–1627
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Markert JA, Champlin DM, Gutjahr-Gobeli R, Grear JS, Kuhn A et al (2010) Population genetic diversity and fitness in multiple environments. BMC Evol Biol 10:205
Meglécz E, Costedoat C, Dubut V, Gilles A, Malausa T, Pech N, Martin JF (2010) QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 26:403–404
Menkhorst P, Richards J (2008) Perameles gunnii. In: IUCN 2010. IUCN Red List of threatened species. Version 2010.1. <www.iucnredlist.org>
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 10:1509–1518
Mitrovski P, Heinze DA, Guthridge K, Weeks AR (2005) Isolation and characterization of microsatellite loci from the Australian endemic mountain pygmy-possum, Burramys parvus broom. Mol Ecol Notes 5:395–397
Myroniuk P (1993) Eastern barred bandicoot recovery. Newsletter no. 3. Conservation and Natural Resources, Victoria
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Robinson NA (1995) Implications from mitochondrial DNA for management to conserve the eastern barred bandicoot (Perameles gunnii). Conserv Biol 9:114–125
Robinson NA, Murray ND, Sherwin WB (1993) VNTR loci reveal differentiation between and structure within populations of the Eastern Barred Bandicoot Perameles gunnii. Mol Ecol 2:195–207
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research. W.H., Freeman, New York
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mor Biol Evol 24:1596–1599
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
DSE Victoria (2009) Eastern barred bandicoot (mainland) Perameles gunni (unnamed subspecies). Action statement no. 4 (revised 2009). Department of Sustainability and Environment, Victoria
Weeks AR, Sgro CM, Young AG, Frankham R, Mitchell NJ et al (2011) Assessing the benefits and risks of translocation in changing environments: a genetic perspective. Evol Appl 4:709–725
Weir BS, Cockerham C (1984) Estimating F statistics for the analysis of population structure. Evolution 38:1358–1370
Willi Y, van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–478
Winnard AL, Coulson G (2008) Sixteen years of Eastern Barred Bandicoot Perameles gunnii reintroductions in Victoria: a review. Pacific Conserv Biol 14:34–53
Zenger KR, Johnston PG (2001) Isolation and characterization of microsatellite loci in the southern brown bandicoot (Isoodon obesulus), and their applicability to other marsupial species. Mol Ecol Notes 1:141–151
Acknowledgments
We thank Megan Lutton and John Roberts for assistance with genotyping, Neil Murray and Peter Courtney for providing historic samples from the last wild population and the captive breeding program, and the eastern barred bandicoot recovery team for discussions. Funding was partially provided by the Department of Sustainability and Environment Victoria and Zoos Victoria for the development of the new microsatellite markers. ARW was funded by the Australian Research Council via their ARC Research Fellowship program. Approval for hair sampling and DNA analysis was given under project AEC 05186.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Weeks, A.R., van Rooyen, A., Mitrovski, P. et al. A species in decline: genetic diversity and conservation of the Victorian eastern barred bandicoot, Perameles gunnii . Conserv Genet 14, 1243–1254 (2013). https://doi.org/10.1007/s10592-013-0512-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0512-9