Abstract.
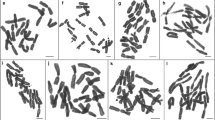
The chromosomes of 31 species of Passiflora, distributed throughout the subgenera Astrophea, Calopathanthus, Distephana, Dysosmia, Passiflora, Plectostemma and Tacsonia were analysed. Three different karyotypes were observed: 2n = 12, 24, 36; 2n = 18, 72 and 2n = 20. The karyotype of these species was almost always constituted of metacentric and submetacentric chromosomes with variable karyotype symmetry. In the group with x = 6, represented by the subgenus Plectostemma, six diploid species with 2n = 12, one tetraploid with 2n = 24 (P. suberosa) and an intraspecific polyploid with 2n = 12, 36 (P. misera) were analysed. P. pentagona (subgenus Astrophea) may also be included in this karyological group since it presents 2n = 24 and may be of polyploid origin, with x = 6. The interphase nuclei in this group were areticulate, except those of P. morifolia and P. pentagona with semi-reticulate characteristics. Two small terminal heterochromatic blocks, positive for chromomycin A3, were identified in the largest chromosome pair of P. capsularis and P. rubra, species very closely related, while P. tricuspis displayed four chromosomes with proximal blocks. In the group with x = 9, represented mainly by subgenus Passiflora, 20 species with 2n = 18 and one with 2n = 72 were studied. They presented chromosomes larger than those species with x = 6 and interphase nuclei of semi-reticulate type, except for P. mixta with areticulate nuclei. Four terminal CMA+ blocks were observed in P. edulis, six blocks in P. caerulea and P. racemosa, while five blocks were observed in the single P. amethystina plant analysed. P. foetida (subgenus Dysosmia), the only species with 2n = 20, exhibited six chromosomes with CMA+ blocks and interphase nuclei of the areticulate type. The meiotic analysis of representatives of the three groups (P. foetida, P. suberosa, P. cincinnata and P. racemosa) always presented regular pairing and regular chromosome segregation, except in P. jilekii where a tetravalent was observed. The analysis of the chromosome variation within the genus and the family suggests that the base number of Passiflora may be x1 = 6 or x1 = 12, whereas x2 = 9 is only an important secondary base number.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received April 11, 2000 Accepted October 5, 2000
Rights and permissions
About this article
Cite this article
De Melo, N., Cervi, A. & Guerra, M. Karyology and cytotaxonomy of the genus Passiflora L. (Passifloraceae). Plant Syst. Evol. 226, 69–84 (2001). https://doi.org/10.1007/s006060170074
Issue Date:
DOI: https://doi.org/10.1007/s006060170074