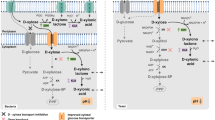
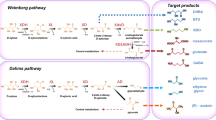
Abstract
The goal of sustainable production of biochemicals and biofuels has driven the engineering of microbial cell as factories that convert low-value substrates to high-value products. Xylose is the second most abundant sugar substrate in lignocellulosic hydrolysates. We analyzed the mechanisms of xylose metabolism using genome sequencing data of 492 industrially relevant bacterial species in the mini-review. The analysis revealed the xylose isomerase and Weimberg pathways as the major routes across diverse routes of bacterial xylose metabolism. In addition, we discuss recent developments in metabolic engineering of xylose metabolism in industrial microorganisms. Genome-scale analyses have revealed xylose pathway-specific flux landscapes. Overall, a comprehensive understanding of bacterial xylose metabolism could be useful for the feasible development of microbial cell factories.



Similar content being viewed by others
References
Alkim C, Trichez D, Cam Y, Spina L, François JM, Walther T (2016) The synthetic xylulose-1 phosphate pathway increases production of glycolic acid from xylose-rich sugar mixtures. Biotechnol Biofuels 9(1):201
Bogorad IW, Lin T-S, Liao JC (2013) Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502:693–697
Bond JQ, Alonso DM, Wang D, West RM, Dumesic JA (2010) Integrated catalytic conversion of gamma-valerolactone to liquid alkenes for transportation fuels. Science 327(5969):1110–1114
Brüsseler C, Radek A, Tenhaef N, Krumbach K, Noack S, Marienhagen J (2017) The myo-inositol/proton symporter IolT1 contributes to d-xylose uptake in Corynebacterium glutamicum. Bioresour Technol 249:953–961
Buschke N, Becker J, Schafer R, Kiefer P, Biedendieck R, Wittmann C (2013) Systems metabolic engineering of xylose-utilizing Corynebacterium glutamicum for production of 1,5-diaminopentane. Biotechnol J 8(5):557–570
Buschke N, Schröder H, Wittmann C (2011) Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J 6(3):306–317
Cabulong RB, Lee W-K, Bañares AB, Ramos KRM, Nisola GM, Valdehuesa KNG, Chung W-J (2018) Engineering Escherichia coli for glycolic acid production from D-xylose through the Dahms pathway and glyoxylate bypass. Appl Microbiol Biotechnol 102(5):2179–2189
Cam Y, Alkim C, Trichez D, Trebosc V, Vax A, Bartolo F, Besse P, François JM, Walther T (2016) Engineering of a synthetic metabolic pathway for the assimilation of (d)-xylose into value-added chemicals. ACS Synth Biol 5(7):607–618
Choi SY, Kim WJ, Yu SJ, Park SJ, Im SG, Lee SY (2017) Engineering the xylose-catabolizing Dahms pathway for production of poly(d-lactate-co-glycolate) and poly(d-lactate-co-glycolate-co-d-2-hydroxybutyrate) in Escherichia coli. Microb Biotechnol 10(6):1353–1364
Choi SY, Park SJ, Kim WJ, Yang JE, Lee H, Shin J, Lee SY (2016) One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat Biotechnol 34(4):435–440
Choi YJ, Lee SY (2013) Microbial production of short-chain alkanes. Nature 502(7472):571–574
Chomvong K, Bauer S, Benjamin DI, Li X, Nomura DK, Cate JH (2016) Bypassing the pentose phosphate pathway: towards modular utilization of xylose. PLoS One 11(6):e0158111
Cirino PC, Chin JW, Ingram LO (2006) Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol Bioeng 95(6):1167–1176
de Vries W, Stouthamer AH (1968) Fermentation of glucose, lactose, galactose, mannitol, and xylose by bifidobacteria. J Bacteriol 96(2):472–478
Dhar KS, Wendisch VF, Nampoothiri KM (2016) Engineering of Corynebacterium glutamicum for xylitol production from lignocellulosic pentose sugars. J Biotechnol 230:63–71
Díaz-Fernández D, Lozano-Martínez P, Buey RM, Revuelta JL, Jiménez A (2017) Utilization of xylose by engineered strains of Ashbya gossypii for the production of microbial oils. Biotechnol Biofuels 10(1):3
Doten RC, Mortlock RP (1985) Inducible xylitol dehydrogenases in enteric bacteria. J Bacteriol 162(2):845–848
El-Semman IE, Karlsson FH, Shoaie S, Nookaew I, Soliman TH, Nielsen J (2014) Genome-scale metabolic reconstructions of Bifidobacterium adolescentis L2-32 and Faecalibacterium prausnitzii A2-165 and their interaction. BMC Syst Biol 8:41
Henard CA, Freed EF, Guarnieri MT (2015) Phosphoketolase pathway engineering for carbon-efficient biocatalysis. Curr Opin Biotechnol 36:183–188
Henry CS, Zinner JF, Cohoon MP, Stevens RL (2009) iBsu1103: a new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol 10(6):R69
Janssen HJ, Steinbuchel A (2014) Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels 7(1):7
Jeffries TW (2006) Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17(3):320–326
Jo S, Yoon J, Lee S-M, Um Y, Han SO, Woo HM (2017) Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J Biotechnol 258:69–78
Jojima T, Noburyu R, Sasaki M, Tajima T, Suda M, Yukawa H, Inui M (2015) Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99(3):1165–1172
Jojima T, Omumasaba CA, Inui M, Yukawa H (2009) Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl Microbiol Biotechnol 85(3):471–480
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104(1–3):5–25
Kang M-K, Lee J, Um Y, Lee TS, Bott M, Park SJ, Woo HM (2014) Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicum and its application to xylose utilization. Appl Microbiol Biotechnol 98(13):1–12
Kawaguchi H, Vertès AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72(5):3418–3428
Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martinez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velazquez-Ramirez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD (2017) The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45(D1):D543–D550
Kim D, Seo SW, Gao Y, Nam H, Guzman GI, Cho BK, Palsson BO (2018) Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res 46(6):2901–2917
Kim SR, Ha S-J, Wei N, Oh EJ, Jin Y-S (2012) Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends Biotechnol 30(5):274–282
Kjeldsen KR, Nielsen J (2009) In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network. Biotechnol Bioeng 102(2):583–597
Lin PP, Jaeger AJ, Wu TY, Xu SC, Lee AS, Gao F, Chen PW, Liao JC (2018) Construction and evolution of an Escherichia coli strain relying on nonoxidative glycolysis for sugar catabolism. Proc Natl Acad Sci U S A 115(14):3538–3546
Liu L, Zhang L, Tang W, Gu Y, Hua Q, Yang S, Jiang W, Yang C (2012) Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194(19):5413–5422
Luterbacher JS, Rand JM, Alonso DM, Han J, Youngquist JT, Maravelias CT, Pfleger BF, Dumesic JA (2014) Nonenzymatic sugar production from biomass using biomass-derived γ-valerolactone. Science 343(6168):277–280
McMillan JD, Jennings EW, Mohagheghi A, Zuccarello M (2011) Comparative performance of precommercial cellulases hydrolyzing pretreated corn stover. Biotechnol Biofuels 4(1):29
Meiswinkel TM, Gopinath V, Lindner SN, Nampoothiri KM, Wendisch VF (2013) Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol 6(2):131–140
Nielsen J, Keasling Jay D (2016) Engineering cellular metabolism. Cell 164(6):1185–1197
Okano K, Yoshida S, Yamada R, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) Improved production of homo-D-lactic acid via xylose fermentation by introduction of xylose assimilation genes and redirection of the phosphoketolase pathway to the pentose phosphate pathway in L-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl Environ Microbiol 75(24):7858–7861
Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BO (2011) A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol Syst Biol 7:535
Pereira B, Li Z-J, De Mey M, Lim CG, Zhang H, Hoeltgen C, Stephanopoulos G (2016) Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab Eng 34:80–87
Radek A, Krumbach K, Gätgens J, Wendisch V, Wiechert W, Bott M, Noack S, Marienhagen J (2014) Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J Biotechnol 192 Pt A:156–160
Radek A, Müller M-F, Gätgens J, Eggeling L, Krumbach K, Marienhagen J, Noack S (2016) Formation of xylitol and xylitol-5-phosphate and its impact on growth of d-xylose-utilizing Corynebacterium glutamicum strains. J Biotechnol 231:160–166
Radek A, Tenhaef N, Müller M-F, Brüsseler C, Wiechert W, Marienhagen J, Polen T, Noack S (2017) Miniaturized and automated adaptive laboratory evolution: evolving Corynebacterium glutamicum towards an improved d-xylose utilization. Bioresour Technol 245:1377–1385
Rossoni L, Carr R, Baxter S, Cortis R, Thorpe T, Eastham G, Stephens G (2018) Engineering Escherichia coli to grow constitutively on D-xylose using the carbon-efficient Weimberg pathway. Microbiology 164(3):287–298
Sasaki M, Jojima T, Inui M, Yukawa H (2010) Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 86(4):1057–1066
Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H (2009) Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol 85(1):105–115
Seo SW, Gao Y, Kim D, Szubin R, Yang J, Cho BK, Palsson BO (2017) Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci Rep 7(1):2181
Seo SW, Kim D, Latif H, O'Brien EJ, Szubin R, Palsson BO (2014) Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910
Seo SW, Kim D, O'Brien EJ, Szubin R, Palsson BO (2015a) Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun 6:7970
Seo SW, Kim D, Szubin R, Palsson BO (2015b) Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep 12(8):1289–1299
Sonderegger M, Schumperli M, Sauer U (2004) Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae. Appl Environ Microbiol 70(5):2892–2897
Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463(7280):559–562
Stephen Dahms A (1974) 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation. Biochem Biophys Res Commun 60(4):1433–1439
Teusink B, Wiersma A, Molenaar D, Francke C, de Vos WM, Siezen RJ, Smid EJ (2006) Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem 281(52):40041–40048
Wang J, Shen X, Lin Y, Chen Z, Yang Y, Yuan Q, Yan Y (2018) Investigation of the synergetic effect of xylose metabolic pathways on the production of Glutaric acid. ACS Synth Biol 7(1):24–29
Wasserstrom L, Portugal-Nunes D, Almqvist H, Sandström AG, Lidén G, Gorwa-Grauslund MF (2018) Exploring D-xylose oxidation in Saccharomyces cerevisiae through the Weimberg pathway. AMB Express 8(1):33
Weimberg R (1961) Pentose oxidation by Pseudomonas fragi. J Biol Chem 236:629–635
Yim SS, Choi JW, Lee SH, Jeon EJ, Chung W-J, Jeong KJ (2017) Engineering of Corynebacterium glutamicum for consolidated conversion of hemicellulosic biomass into xylonic acid. Biotechnol J 12(11):1700040
Yim SS, Choi JW, Lee SH, Jeong KJ (2016) Modular optimization of a hemicellulose-utilizing pathway in Corynebacterium glutamicum for consolidated bioprocessing of hemicellulosic biomass. ACS Synth Biol 5(4):334–343
Zhang G-C, Liu J-J, Kong II, Kwak S, Jin Y-S (2015) Combining C6 and C5 sugar metabolism for enhancing microbial bioconversion. Curr Opin Chem Biol 29:49–57
Zhang J, Babtie A, Stephanopoulos G (2012) Metabolic engineering: enabling technology of a bio-based economy. Curr Opin Chem Eng 1(4):355–362
Zhu X, Zhao D, Qiu H, Fan F, Man S, Bi C, Zhang X (2017) The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway. Metab Eng 43:37–45
Acknowledgements
The authors appreciate fruitful discussions with Mr. Jungseok Lee, Ms. Suah Jo, and Mr. Seung Soo Lee. Also, the authors appreciate helpful discussions with Ms. Ina Bang.
Funding
This work was supported by Korea CCS R&D Center (KCRC) (2017M1A8A1072034) and Basic Science Research Program (2017R1A2B2002566) through the National Research Foundation of Korea, funded by the Korean Government (Ministry of Science and ICT). In addition, this work was partially supported by the Golden Seed Project (213008-05-2-WT911) grant, funded by the Ministry of Agriculture and the Ministry of Oceans and Fisheries. Financial support from the CJ Grant Program (CG-20-16-01-0003) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not involve any studies with human participants performed by any of the authors.
Consent for publication
Not applicable.
Electronic supplementary material
ESM 1
(PDF 24 kb)
Rights and permissions
About this article
Cite this article
Kim, D., Woo, H.M. Deciphering bacterial xylose metabolism and metabolic engineering of industrial microorganisms for use as efficient microbial cell factories. Appl Microbiol Biotechnol 102, 9471–9480 (2018). https://doi.org/10.1007/s00253-018-9353-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9353-2