Abstract
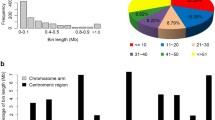
Introgression lines (ILs) are useful tools for precise mapping of quantitative trait loci (QTLs) and the evaluation of gene action or interaction in theoretical studies. A set of 159 ILs carrying variant introgressed segments from Chinese common wild rice (Oryza rufipogon Griff.), collected from Dongxiang county, Jiangxi Province, in the background of Indica cultivar (Oryza sativa L.), Guichao 2, was developed using 126 polymorphic simple sequence repeats (SSR) loci. The 159 ILs represented 67.5% of the genome of O. rufipogon. All the ILs have the proportions of the recurrent parent ranging from 92.4 to 99.9%, with an average of 97.4%. The average proportion of the donor genome for the BC4F4 population was about 2.2%. The mean numbers of homozygous and heterozygous donor segments were 2 (ranging 0–8) and 1 (ranging 0–7), respectively, and the majority of these segments had sizes less than 10 cM. QTL analysis was conducted based on evaluation of yield-related traits of the 159 ILs at two sites, in Beijing and Hainan. For 6 out of 17 QTLs identified at two sites corresponding to three traits (panicles per plant, grains per panicle and filled grains per plant, respectively), the QTLs derived from O. rufipogon were usually associated with an improvement of the target trait, although the overall phenotypic characters of O. rufipogon were inferior to that of the recurrent parent. Of the 17 QTLs, 5 specific QTLs strongly associated with more than one trait were observed. Further analysis of the high-yielding and low-yielding ILs revealed that the high-yielding ILs contained relatively less introgressed segments than the low-yielding ILs, and that the yield increase or decrease was mainly due to the number of grain. On the other hand, low-yielding ILs contained more negative QTLs or disharmonious interactions between QTLs which masked trait-enchancing QTLs. These ILs will be useful in identifying the traits of yield, tolerance to low temperature and drought stress, and detecting favorable genes of common wild rice.




Similar content being viewed by others
References
Aggarwal RK, Brar DS, Khush GS (1997) Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254:1–12
Alpert K, Tanksley SD (1996) High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit-weight quantitative trait locus in tomato. Proc Natl Acad Sci USA 93:15503–15507
van Berloo R (1999) Computer note. GGT: software for the display of graphical genotypes. J Hered 90:328–329
Bernacchi D, Beck-Bunn T, Eshed Y, Inai S, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S (1998a) Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397
Bernacchi D, Beck-Bunn T, Emmatty D, Eshed Y, Inai S, Lopez J, Petiard V, Sayama H, Uhlig J, Zamir D, Tanksley S (1998b) Advanced back-cross QTL analysis of tomato. II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon hirsutum and L. pimpinellifolium.. Theor Appl Genet 97:170–180
Brondani C, Rangel PHN, Brondani RPV, Ferreira ME (2002) QTL mapping and introgression of yield-related traits from Oryza glumaepatula to cultivated rice (Oryza sativa) using microsatellite markers. Theor Appl Genet 104:1192–1203
Cai HW, Morishima H (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet 104:1217–1228
Chetelat RT, Meglic V (2000) Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor Appl Genet 100:232–241
Chetelat RT, De Verna JW, Bennett AB (1995) Introgression into tomato (Lycopersicon esculentum) of the L. chmielewskii sucrose accumulator gene (sucr) controlling fruit sugar composition. Theor Appl Genet 91:327–333
Clayton WD, Renvoize SA (1986) Genera graminum. Grasses of the world. Kew Bulletin Series VIII. Her Majesty’s Stationary Office, London
Coffman WR, Herrera RM (1980) Rice. In: Fehr WR, Hadley HH (eds) Hybridization of crop plants. American Society of Agronomy-Crop Science of America, Madison, pp 101–131
Doi K, Iwata N, Yoshimura A (1997) The construction of chromosome introgression lines of African rice (Oryza glaberrima Steud.) in the background of japonica (O. sativa L.). Rice Genet Newsl 14:39–41
Eshed Y, Zamir D (1994) Introgressions from Lycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theor Appl Genet 88:891–897
Eshed Y, Zamir D (1995) Introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield associated QTL. Genetics 141:1147–1162
Eshed Y, Zamir D (1996) Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143:1807–1817
Grandillo S, Ku HM, Tanksley SD (1996) Characterization of fs8.1, a major QTL influencing fruit shape in tomato. Mol Breed 2:251–260
Howell PM, Marshall DF, Lydiate DJ (1996) Towards developing inter-varietal introgression lines in Brassica napus using marker-assisted selection. Genome 39:348–358
Khush GS, Ling KC, Aguiero VM (1977) Breeding for resistance to grassy stunt in rice. In: Proceedings of the 3rd international congress SABRAO. Canberra, Australia. Plant Breed, Paper I, pp 3–9
Khush GS, Bacalangco E, Ogawa T (1990) A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet Newsl 7:121–122
Koumproglou R, Wilkes TM, Townson P, Wang XY, Beynon J, Pooni HS, Newbury HJ, Kearsey MJ (2002) STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J 31:355–364
Kubo T, Aida Y, Nakamura K, Tsunematsu H, Doi K, Yoshimura A (2002) Reciprocal chromosome segment substitution series derived from Japonica and Indica cross of rice (Oryza sativa L.). Breed Sci 52:319–325
Li DJ, Sun CQ, Fu YC, Chen L, Zhu ZF, Li C, Cai HW, Wang XK (2002) Identification and mapping of genes for improving yield from Chinese common wild rice (O. rufipogon Griff.) using advanced backcross QTL analysis. Chin Sci Bull 18:1533–1537
Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Matus I, Corey A, Filichkin T, Hayes PM, Vales MI, Kling J, Riera-Lizarazu O, Sato K, Powell W, Waugh R (2003) Development and characterization of recombinant chromosome introgression lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Genome 46:1010–1023
Moncada P, Martinez CP, Borrero J, Chatel M, Gauch H, Guimaraes E, Tohme J, McCouch SR (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–52
Monforte AJ, Tanksley SD (2000) Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor Appl Genet 100:471–479
Oka HI (1988) Origin of cultivated rice. Developments in crop science, vol 14. Elsevier, Amsterdam
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Paterson AH, Deverna JW, Lanini B, Tanksley SD (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes in an interspecies cross of tomato. Genetics 124:735–742
Rangel PH, Guimarães EP, Neves PCF (1996) Base genética das cultivares de arroz (Oryza sativa L.) irrigado do Brasil. Pesqui Agropec Bras 31:349–357
Rogers OS, Bendich AJ (1988) Extraction of DNA from plant tissue, plant molecular. Biol Manual A6:1–10
Second G (1982) Origin of the genetic diversity of cultivated rice (Oryza spp.), study of the polymorphism scored at 40 isozyme loci. Jpn J Genet 57:25–57
Septiningsih EM, Prasetiyono J, Lubis E, Tai TH, Tjubaryat T, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1419–1432
Sobrizal, Ikeda K, Sanchez PL, Doi K, Angles ER, Khush GS, Yoshimura A (1999) Development of Oryza glumaepatula introgression line in rice, O. sativa L. Rice Genet Newsl 16:107–108
Sun CQ, Wang XK, Yoshimura A, Iwata N (2001) Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor Appl Genet 102:157–162
Sun CQ, Wang XK, Yoshimura A, Doi K (2002) Genetic differentiation for muclear, mitochondrial and chloroplast genomes in common wild rice (O. rufipogon Griff.) and cultivated rice (O. sativa L.). Theor Appl Genet 104:1335–1345
Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subnit of protein kinase CK2. Proc Natl Acad Sci USA 98:7922–7927
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Temnykh S, Park W, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinea CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet 83:565–581
Xiao J, Grandillo S, Ahn SN, McCouch SR, Tanksley SD, Li J, Yuan L (1996) Genes from wild rice improve yield. Nature 384:223–224
Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150:899–909
Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M (1998) Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factor. Theor Appl Genet 97:37–44
Yamamoto T, Lin HY, Sasaki T, Yano M (2000) Identification of heading date quantitative trait loci Hd6 and characterization of its epistatic interaction with Hd2 in rice using advanced backcross progeny. Genetics 154:885–891
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Young ND, Tanksley SD (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet 77:95–101
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989
Zhang Q, Zhao BY, Zhao KJ, Wang CL, Yang WC, Lin SC, Jue GS, Zhou YL, Li DY, Chen CB, Zhu LH (2000) Identification and mapping a new gene Xa-23(t) for resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) from O.rufipogon. Acta Agron Sin 26:536–542
Acknowledgements
We thank two anonymous reviewers for their valuable comments on the manuscript. This work was funded by the National Natural Science Foundation of China (No. 30270803), the “973” Project of the Ministry of Sciences and Technology of China (No. 2001CB108800), a grant from China National High-Tech Research and Development (“863”) Program (No. 2003AA207040), and a grand from Protection and Utilization of Agricultural Wild Plants Program of the Ministry of Agriculture of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Sasaki
Rights and permissions
About this article
Cite this article
Tian, F., Li, D.J., Fu, Q. et al. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet 112, 570–580 (2006). https://doi.org/10.1007/s00122-005-0165-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0165-2