Abstract
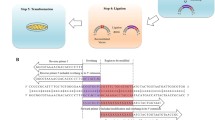
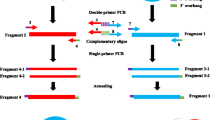
Most DNA shuffling methods currently used require PCR process. A novel method of DNA shuffling without PCR process is described, taking advantage of the feature of some restriction enzymes whose recognition sites differ from their cleavage sites, thus giving rise to different cohesive ends. These cohesive ends can be rejoined at their native sites from different parental sequences, generating new sequences with various combinations of mutations.
Similar content being viewed by others
References
Stemmer, W. P., DNA shuffling by random fragmentation and reassembly:in vitro recombination for molecular evolution, Proc. Natl. Acad. Sci. USA, 1994, 91(22): 10747–10751.
Stemmer, W. P., Rapid evolution of a proteinin vitro by DNA shuffling, Nature, 1994, 370(6488): 389–391.[DOI]
Zhao, H., Arnold, F. H., Optimization of DNA shuffling for high fidelity recombination, Nucleic Acids Res., 1997, 25(6): 1307–1308.[DOI]
Zhao, H., Giver, L., Shao, Z. et al., Molecular evolution by staggered extension process (StEP)in vitro recombination, Nat. Biotechnol., 1998, 16(3): 258–261.
Shao, Z., Zhao, H., Giver, L. et al., Random-primingin vitro recombination: an effective tool for directed evolution, Nucleic Acids Res., 1998, 26(2): 681–683.[DOI]
Crameri, A., Raillard, S. A., Bermudez, E. et al., DNA shuffling of a family of genes from diverse species accelerates directed evolution, Nature, 1998, 391(6664): 288–291.[DOI]
Kikuchi, M., Ohnishi, K., Harayama, S., Novel family shuffling methods for thein vitro evolution of enzymes, Gene, 1999, 236(1): 159–167.[DOI]
Kikuchi, M., Ohnishi, K., Harayama, S., An effective family shuffling method using single-stranded DNA, Gene, 2000, 243(1–2): 133–137.[DOI]
Ness, J. E., Kim, S., Gottman, A. et al., Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently, Nat. Biotechnol., 2002, 20(12): 1251–1255.[DOI]
Wang, Q., Liu, Q., Li, B., Simultaneous detection of 7 mutations with 7 forward primers and 1 common reverse primer in a single PCR step. J Biochem Biophys Methods, 2004, 58(2): 153–157.[DOI]
Huang, M. M., Arnheim, N., Goodman, M. F., Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR, Nucleic Acids Res., 1992, 20(17): 4567–4573.
Kwok, S., Kellogg, D. E., McKinney, N. et al., Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies, Nucleic Acids Res., 1990, 18(4): 999–1005.
Sarkar, G., Cassady, J., Bottema, C. D. K. et al., Characterization of polymerase chain reaction amplification of specific alleles, Anal. Biochem., 1990, 186(1): 64–68.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Wang, Q., Liu, Q., Li, G. et al. A novel method of DNA shuffling without PCR process. Chin.Sci.Bull. 49, 689–691 (2004). https://doi.org/10.1007/BF03184266
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03184266